Abstract
Objectives
Brain neoplasms or intracranial tumors, which are more common in older adults, can affect individuals of any age including pediatric and children. Exposure to carcinogenic agents including ionizing radiation and family history is one of the main causes of the disease. Early diagnosis is crucial to avoid prolonged. patients' suffering. The aim of the study was to efficiently recognize the brain tumors from the other brain tissues which include grey and white matter as well as cerebrospinal fluid (CSF).
Materials and methods
This study was performed using axial, sagittal and coronal views for fifty brain tumor patients randomly selected from a set of 200 patients, with a “control” set consisting of images showing no sign of disease; and the “test” brain MRI images for patients diagnosed with brain tumor. The study includes both genders with age ranging from 18 years to 83 years old, (56.5 ± 17.2). The brain images were acquired using a standard head coil Philips Intera 1.5 Tesla machine (USA). The thickness of each section in the entire sequence was 8 mm. Acquisition of T2-weighted and T1-weighted were performed. Interactive Data Language software was used to analyze the data.
Results
The results of this study showed that: the overall accuracy of classification process was 94.8%, and for the tumor; the sensitivity was 97.3%. White matter and grey matter showed a classification accuracy of 95.7% and 89.7% and for CSF the accuracy was 94.3%.
Conclusion
The results showed that brain tumor can be classified successfully and delineated using texture analysis with minimum efforts and with high accuracy for brain tumors.
Keywords: Brain tumors, MRI, Texture analysis, Interactive Data Language, T1-T2-Weighted
1. Introduction
Brain neoplasms or intracranial tumors, which is more common in older adults, can affect individuals of any age including pediatric and children. Exposure to carcinogenic agents including ionizing radiation and family history are among the main causes of the disease. Early diagnosis is crucial to avoid prolonged period of morbidity and help in reduction of mortality rate by avoiding life threatening complications and providing effective treatment on time. (Abd-Ellah et al., 2019) When the mechanisms controlling the growth of normal cells fail to function, tumors start growing and multiplying uncontrollably. This uncontrolled tumor cell growth interrupts the normal tissue and creates a shear stress on cells eventually killing them. They occupy the cavities within the skull interrupting the normal functions of brain. This elevates the intracranial pressure and damages the nerves of the healthy tissue, not to mention the severe pain that it creates. The tumor may be malignant or benign. World Health Organization (WHO) classified brain tumor into 120 types based on the degree of aggression in behavior, and whether they are primary or secondary tumors (Louis et al., 2016, Brain tumor society). Benign tumors usually grow slowly and are quite localized. Each of these tumors show unique radiographic and biological characteristics when imaged.
A variety of imaging techniques including magnetic resonance imaging (MRI), computed tomography (CT) scans, positron emission tomography (PET) and single photon emission computed tomography (SPECT) scans, and cerebral angiography are used to study brain tumors. CT and MR imaging are preferred in many centers due to their availability, cost-effectiveness as well as their ability to produce high resolution images describing anatomy and pathology. It is commonly used for brain tumor imaging and has benefits over CT for being: nonionizing, which minimizes the harm to health tissues, highly efficient in terms of generating 3-D images allowing for precise localization of tumors, lower sensitivity to contrast agents, possessing a wider range of soft tissue contrast, and last but not least, capable of acquisition of both functional and anatomical information simultaneously with limited necessity to physically move the patient. Nevertheless, they do not pick up the bones of the skull as well as CT scans and therefore may not show the effects of tumors on the skull (American Cancer Society, 2020a, American Cancer Society, 2020b). There are a few reactions to MRI scanning including: panic, nausea, headache, and pain resulting from the injected contrast material (American Cancer Society, 2020a, American Cancer Society, 2020b).
Several digital image processing and algorithms have been used to view, analyze, and interpret brain images with a goal to make clinically valuable decisions by improving image perception. Texture analysis has been reported since the seventies of last century (Haralick et al., 1973). Image texture is commonly defined as function of the spatial variation of pixel intensities in an image. Using three-dimensional (3D) computer graphics applications it generates highly complex and realistic looking surfaces, which provides quantitative information in the form of texture features than cannot be visualized and discriminated by the human eye. The features used during texture analysis are mathematical parameters calculated from the distribution of pixels, which characterize the texture type in the image. The main image processing disciplines in which texture analysis techniques are used are classification, segmentation, and synthesis of dissimilar features including normal tissue, inflammatory regions and tumors (Pietikäinen et al., 2000, Alves et al., 2020).
The number of pixels used define the local features and accordingly classify them into first-order and second order textures. The properties of individual pixel values, are estimated using first-order textures which, in turn use image histograms that include variance; coarseness; skewness; kurtosis; energy; and entropy. Here, the spatial interaction between the neighboring image pixels. are not taken into account.
Meanwhile, second-order textures estimate properties of several pixel values relative to the neighboring pixels. They are based on calculation of grey tone spatial dependency matrices (GTSDM) elements according to co-occurrence matrix. These include amongst other features: angular second moment, contrast and correlation that can be calculated using different window size.
It was estimated that morality rate was increased up to 300 times as a direct result of brain tumors among developed countries populations with Western and Central Europe had the highest incidence rates, East and South East Asia showed lower incidences (Leece et al., 2017, GBD, 2016). The objective of the imaging modality is to provide clinical finding regarding the tumor existence, size, location, and type. The accurate and prompt diagnosis will usually provide better intervention options for the treatment and increase the survival rate of the patients (Abd-Ellah et al., 2019). Many researches were published regarding brain tumor diagnosis during various image processing techniques to improve tumor identification and precise location determination (Katouli and Rahmani, 2020, Alqudah and Al-Sharu, 2020, Gulo et al., 2019, Naceur et al., 2018, Pandey et al., 2016). However, the challenge of accurate diagnosis still exists because various tumors have different patterns. Also, it is not easy to discriminate between normal and cancerous tissues at early stage. Therefore, any effort helps the clinician in accurate diagnosis is welcome and it will help in saving lives. The aim was to recognize the brain tumors from the other brain tissues which include: grey and white matter as well as cerebrospinal fluid (CSF).
2. Materials and methods
The study presents an analytical study on MRI images collected at Alnelain hospital in Khartoum state, Sudan using Head Coil Philips Intera 1.5 Tesla (USA) MRI machine. It uses normal MRI brain images and MRI images with brain tumors for classification and delineation of tumor boarder. The population of this study was data set (brain MR Images), with a “control” set consisting of images showing no sign of disease; and the “test” brain MRI images for patients diagnosed with brain tumor. The study includes both genders with age ranging from 18 years to 83 years old.
This study was performed using axial, sagittal and coronal views for fifty brain tumor patients randomly selected from a set of 200 patients. The brain images were acquired using a standard head coil Philips Intera 1.5 Tesla machine. The thickness of each section in the entire sequence was 8 mm. Acquisition of T2-weighted and T1-weighted were performed.
The collected images were then viewed using the Radiant, Ant DICOM to select the axial images suitable for the criteria set for the research population before uploading them as TIFF format into the computer-based software Interactive Data Language (IDL): a vector-oriented programing language designed by David Stern that shares a particular syntax with a PV-wave using a codebase (Bowman, 2005). Areas representing: the background, grey matter, white matter, cerebrospinal fluid (CSF) and tumor, in case of test group, were marked. Windows 3 × 3 pixels were generated in the marked areas and textural feature for the classes center were observed. These textural features include signal-to-noise ratio, energy, entropy, standard deviation, coefficient of variation (COV) and variance. These features were assigned as classification center used by the Euclidian distances to classify the whole image. The algorithm scans and computes the textural features studied and then computes the Euclidean distance, between the calculated features during the scanning and the center of the classes. It then assigns the window to the class with the lowest distance. The same processes for other positions was performed over again till the entire image were classified and classification maps were generated.
The data concerning the brain tissues (CSF, grey, and white matter) and tumor were then entered into SPSS to generate a classification score using stepwise linear discriminate analysis. This aims to select the most discriminate features that can be used in the classification of brain tissues in MRI images. Scatter plots using discriminate function were generated. Moreover, classification accuracy and linear discriminate function equations were used to classify the brain tissues into the previous classes without segmentation process for unseen images in routine work. The delineation of brain tumor was done by further processing of the classification using region label function to segment the brain tumor from the other classes and convert the segmented brain tumor from classification map with pseudo-color to binary image to extract the brain tumor from the whole original image. Then by applying Sobel function the outline of the binary image was generated, and the spatial location of the pixels was used to delineate the brain tumor on the original image using read line.
3. Results:
This study aims to classify the brain image in axial MRI images into normal brain tissues (white matter, grey matter, cerebrospinal fluid ‘CSF’) and tumor, using k-means and linear discriminant analysis.
The classification maps shown in Fig. 1 were generated using Euclidean distance. Here, the centers of the classes were chosen arbitrary from areas representing the classes of interest: white matter, grey matter, cerebrospinal fluid, and tumor.
Fig. 1.
Scatter plot generated using discriminant analysis function for four classes representing: white matter, grey matter, CSF and tumor.
Fig. 2 shows: an original image used in the study 2 (A), the classification map of the original image using Euclidian distance 2 (B) an original image with tumor region delineated 2 (C).
Fig. 2.
(A) An original axial MRI image (B) classification map of the original image using Euclidian distance, (C) an original image with tumor region delineated.
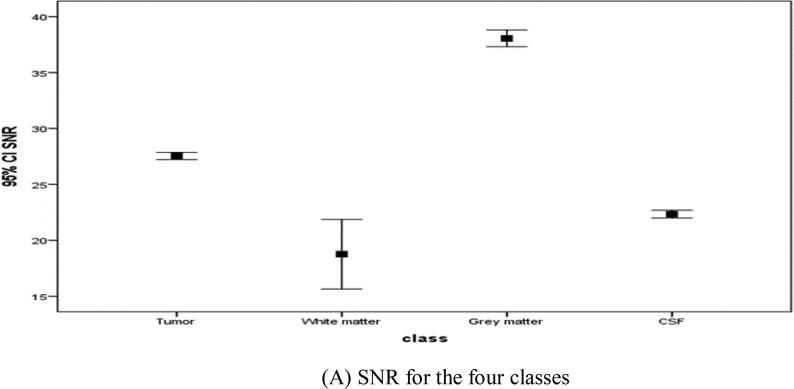
The signal-to-noise ratio and energy graphs were plotted for the different classes, as shown in Fig. 3A, Fig. 3B respectively.
Fig. 3A.
SNR for the four classes.
Fig. 3B.
Energy graphs for the four classes.
Fig. 4: shows error bar plot for the (A) entropy and (B) standard deviation for textural features computed using the linear stepwise discriminant function as a discriminant feature. In 4 (A) it discriminates between gray matter and CSF and in 4 (B) it discriminates between CSF and other features.
Fig. 4.
error bar plot for the (A) entropy and (B) standard deviation.
Fig. 5 shows error bar plot for the (A) COV and (B) variance for textural features computed using linear stepwise discriminant function as a discriminant feature.
Fig. 5.
Error bar plot for the (A) coefficient of variation and (B) Variance.
Figure error bar plot for the (A) coefficient of variation and (B) Variance
4. Discussions
Brain tumors largely reduces the quality of life; both of patients, and their families. They inflict people irrespective of gender, race and ethnicities although it is more associated with elders than infants and teenagers (National Brain Tumor Society). The annual rate of confirmed brain metastases cases range between 70,000 to 170,000 cancer patients and the annual mortality rate is around 100,000 (Workshop on Product Development for Central Nervous System Metastases, 2019). Brain tumors are very difficult to cure leading to high morbidity due to progressive neurologic dysfunction (Seano et al., 2019). One of the most popular modalities for investigating brain tumors is MRI which is often used to visualize pathological or other physiological alterations of living tissues. A major limitation is that only experienced radiologists and clinical experts can segment, detect, and extract infected tumor areas with high accuracy. Computer aided technology helps to overcome this limitation (Bahadure et al., 2017).
The main aim of this study was to classify the brain image in axial MRI images into normal brain tissues: white matter, grey matter, cerebrospinal fluid ‘CSF’, and tumor using k-means and linear discriminant analysis. The study includes both genders with age ranging from 18 years to 83 years old, (56.5 ± 17.2) who underwent MRI brain imaging at Alnelain hospital in Khartoum.
In this study six features: signal-to-noise ratio, energy, entropy, standard deviation, coefficient of variation and variance, were extracted from normal and abnormal (tumor) brain MRI images performed by two experienced radiologists using 3 × 3 window. All patients were informed about the study and how it will not affect the results of the MRI. No IRB was required because all data were originally obtained for diagnostic purposes with the ethical approval from the hospital. All features showed significant correlation with the predefined classes (white matter, grey matter, cerebrospinal fluid, and tumor). Imaging protocols followed was imaging the brain using a standard head coil, with section thickness of 8 mm on all sequences for acquisition of T2-weighted and T1-weighted
The algorithm classified these tissues using the selected centers as initial guess. The algorithm then iterates five cycles, in each cycle it changes the classes centers using the average value of each of the features allocated to the class. The changes in the class center were small fractions which approached zero change in the third iteration. The final designated class centers represent the class of interest Fig. 2(B).
Considering discriminant power of the applied features; the signal-to-noise ratio (SNR) could be differentiated between all the classes successfully Fig. 3A. Moreover, the energy graphs for the different classes Fig. 3B showed similar discriminant power.
From Fig. 4(A) and (B), entropy can successfully differentiate the CSF and gray matter from the rest of the tissues, but it does not show any variation between the other tissues. This is because the white matter has larger standard deviation compared to the average than other tissues. This situation reduces the discriminant power between the tumor and white matter as shown in (Fig. 4A). The standard deviation feature, on the other hand, differentiates well between the CSF and the rest of the tissue, where CSF showed lower variation than other tissues. This is shown clearly in Fig. 5B
From Fig. 5: The coefficient of variation discriminates between the white matter and the rest of the tissue. Fig. 5(A) shows that white matter had inflated value while all tissues showed low COV. Similar results were achieved by the textural variable variance Fig. 5(B).
Tumor and other brain tissues in MRI images, for simplicity, were diagnosed as normal or abnormal by using the following simple equations after extracting the associated features using a window of 3 × 3 pixel from the region of interest; the biggest classification score assumes the tissue type.
| Tumor = (0.602 × COV) + (-0.126 × variance) + (-4.349 × STD) +(17.195 × SNR) + (0.00008 × energy) +(-0.061 × entropy) −109.37 |
| White matter = (0.511 × COV) + (-0.033 × variance) + (-3.106 × STD) + (13.146 × SNR) + (0.0001 × energy) (-0.057 × entropy) + -54.627 |
| Grey matter = (0.496 × COV) + (-0.133 × variance) + (-2.671 × STD) +(12.576 × SNR) + (0.000046 × energy) (-0.040 × entropy) + -74.131 |
| CSF = (0.645 × COV) + (-0.063 × variance) + (-5.739 × STD) +(19.374 × SNR) + (0.00012 × energy) (-0.076 × entropy) −119.124 |
To classify the grey and white matter including cerebrospinal fluid as normal and abnormal (tumor), the raw data representing the features of the classified regions of the whole images were further classified using linear discriminate analysis. Table 1 shows the accuracy levels of the different tissue types.
Table 1.
Classification score matrix generated by linear discriminant analysis.
| Original group | Predicted Group Membership |
Total | ||||
|---|---|---|---|---|---|---|
| Tumor | White matter | Grey matter | CSF | |||
| % |
Tumor | 97.3 | 0 | 0 | 2.7 | 100% |
| White matter | 0 | 95.7 | 2.6 | 1.7 | 100% | |
| Grey matter | 5.1 | 5.1 | 89.7 | 0 | 100% | |
| CSF | 5.7 | 0 | 0 | 94.3 | 100% | |
| Total classification accuracy = 94.8% | ||||||
From Table 1, the result of the classification showed that the tumor areas were successfully distinguished from the rest of the tissues despite its similarity with the surrounding tissue. The classification accuracies of the different tissues were tumor 97.3%, the white matter 95.7, gray matter 89.7%, and CSF 94.3%. The overall accuracy was 94.8%. The parameters studied were similar to Hassan et al. who used texture analysis for glioma using three different windows and whose classification accuracy ranged between 98.1% and 99.5% (Hassan et al., 2016).
The limitation of this study is that it did not test all possible texture analysis to decide the best type. Nor did it study the effect of texture analysis in other imaging modalities. Moreover, the accuracy of detection was comparable to other authors who used other techniques (Kamil and Altaei, 2020, Ouchtati et al., 2019, Bahadure et al., 2017)
5. Conclusion
This study aims to segment and classify the brain MRI images as normal tissues (white matter, grey matter and cerebrospinal fluid) and abnormal tissues (tumor) using texture analysis. multiple features of first order statistics were included: signal-to-noise ratio, energy, entropy, standard deviation, coefficient of variation and variance .The study found that the tumor texture revealed a very different underlying pattern compared to the other tissues of the brain, which allows better allocation than visual assessment. The classification accuracy of the tumor was 97.3%. The accuracy of detecting normal brain tissue was: white matter 95.7, gray matter 89.7%, and CSF 94.3%. The overall accuracy was 94.8% using linear discriminant analysis. The results are quite promising showing that texture analysis can be used with high accuracy in both diagnosis and therapy of brain tumors, were sensitive healthy and very radiosensitive tissue lie at a very close proximity to brain tumors.
6. Recommendations
The ability of texture analysis techniques to recognize different tumors, using other images should be validated using the studied parameters as well as others. If texture analysis showed accurate results in all types of tumors, then it can be implemented, which will ensure better diagnosis and therapy in minimal time.
CRediT authorship contribution statement
Buthayna G.Elshaikh: Conceptualization, Data curation, Formal analysis, Methodology, Software, Supervision, Validation, Visualization, Writing - original draft, Writing. MEM Garelnabib: Conceptualization, Data curation, Formal analysis Methodology, Software, Supervision, Validation, review & editing. Hiba Omer: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Software, Validation, Visualization, Writing - review & editing. Abdelmoneim Sulieman: Conceptualization, Methodology, Project administration, Resources, Software, Supervision, Validation, review & editing. B. Habeeballa: Conceptualization, Data curation, Formal analysis Methodology, Software, Supervision, Validation, Visualization, Writing - original draft. Rania A.Tabeidi: Conceptualization, Data curation, Formal analysis Methodology, Software, Validation, Visualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abd-Ellah M.K., Awad A.I., Khalaf A.A.M., Hamed H.F.A. A review on brain tumor diagnosis from MRI images: Practical implications, key achievements, and lessons learned. Magn Reson Imaging. 2019;61:300–318. doi: 10.1016/j.mri.2019.05.028. [DOI] [PubMed] [Google Scholar]
- Alqudah A., Al-Sharu W. Brain tumor classification using deep learning technique–A comparison between cropped, uncropped, and segmented lesion images with different sizes. Image Video Processing (eess IV) 2020;8(6):3684–3691. [Google Scholar]
- Alves A., Miranda J., Reis F., de Souza S., Alves L., Feitoza L., de Castro J., de Pina D. Inflammatory lesions and brain tumors: Is it possible to differentiate them based on texture features in magnetic resonance imaging? J. Venom Anim. Toxins Incl. Trop. Dis. 2020;26:1–10. doi: 10.1590/1678-9199-JVATITD-2020-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Societ, 2020. Tests for Brain and Spinal Cord Tumors in Adults available at https://www.cancer.org/cancer/brain-spinal-cord-tumors-adults/detection-diagnosis-staging/how-diagnosed.html.
- American Cancer Society 2020, MRI for cancer. available at: https://www.cancer.org/treatment/understanding-your-diagnosis/tests/mri-for-cancer.html.
- Bahadure N., Ray A., Thethi H. Image Analysis for MRI Based Brain Tumor Detection and Feature Extraction Using Biologically Inspired BWT and SVM. Int. J. Biomed. Imaging. 2017;2017:1–12. doi: 10.1155/2017/9749108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain tumor society, Found at: https://braintumor.org/brain-tumor-information/brain-tumor-facts/
- GBD 2016 Brain and Other CNS Cancer Collaborators. Global, regional, and national burden of brain and other CNS cancer, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019 Apr;18(4):376-393. [DOI] [PMC free article] [PubMed]
- Bowman K. An Introduction to Programming with IDL: Interactive Data Language. Academic Press. 2005:1–312. [Google Scholar]
- Gulo C., Sementille A., Tavares J. Techniques of medical image processing and analysis accelerated by high-performance computing: A systematic literature review. J. Real-Time Image Proc. 2019;16(6):1891–1908. [Google Scholar]
- Haralick M., Shanmugam K., Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybernet. 1973;3(6):610–621. [Google Scholar]
- Hassan A., Gar-elnabi M., Taha E., Ahmed A., Awad A.B. Characterization of Brain Glioma in MRI using Image Texture Analysis Techniques. Int. J. Sci. Res. 2016;5(3):817–821. [Google Scholar]
- Kamil S., Altaei M. Brain Tumor Diagnosis Using MR Image Processing. Al-Nahrain Journal of Science. 2020;23(2):67–74. [Google Scholar]
- Katouli M., Rahmani A. Brain Tumor Diagnosis in MRI Images Using Image Processing Techniques and Pixel-Based Clustering. Traitement du Signal. 2020;37(2):291–300. [Google Scholar]
- Leece R., Xu J., Ostrom Q., Chen Y., Kruchko C., S Barnholtz-Sloan J. Global incidence of malignant brain and other central nervous system tumors by histology, 2003–2007 Neuro Oncol. 2017; 19(11): 1553–1564 [DOI] [PMC free article] [PubMed]
- Louis D., Perry A., von Reifenberger G., Deimling A., Figarella-Branger D., Cavenee W., Ohgaki H., Wiestler O., Kleihues P., Ellison D. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Naceur M.B., Saouli R., Akil M., Kachouri R. Fully Automatic Brain Tumor Segmentation using End-To-End Incremental Deep Neural Networks in MRI images. Comput. Meth. Prog. Biomed. 2018 Nov;166:39–49. doi: 10.1016/j.cmpb.2018.09.007. [DOI] [PubMed] [Google Scholar]
- National Brain Tumor Society. Quick Brain Tumor Facts. Available at: https://braintumor.org/brain-tumor-information/brain-tumor-facts/.
- Ouchtati S., Chergui A., Mavromatis S., Belmeguenai A., Rafik D., Sequeira J. Novel Method for Brain Tumor Classification Based on Use of Image Entropy and Seven Hu’s Invariant Moments. Traitement du Signal, Lavoisier. 2019;36(6):483–491. [Google Scholar]
- Pandey A., Beg S., Yadav A. Image processing technique for the enhancement of brain tumor pattern. J. Res. Develop. Appl. Sci. Eng. 2016;9(2):1–4. [2] [Google Scholar]
- Pietikäinen M., Ojala T., Xu Z. Rotation-invariant texture classification using feature distributions. Pattern Recogn. 2000;33(1):43–52. [Google Scholar]
- Seano G., Nia H., Emblem K., Datta M., Ren J., Krishnan S. Solid stress in brain tumors causes neuronal loss and neurological dysfunction and can be reversed by lithium. Nat. Biomed. Eng. 2019;3(3):230–245. doi: 10.1038/s41551-018-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workshop on Product Development for Central Nervous System Metastases, 2019. available at https://braintumor.org/wp-content/assets/Brain-Mets-Workshop-Summary-2019-FINAL.pdf.