Figure 1.
Experimental Analysis of CpG-Rich Sequences in a Gene-Desert BAC
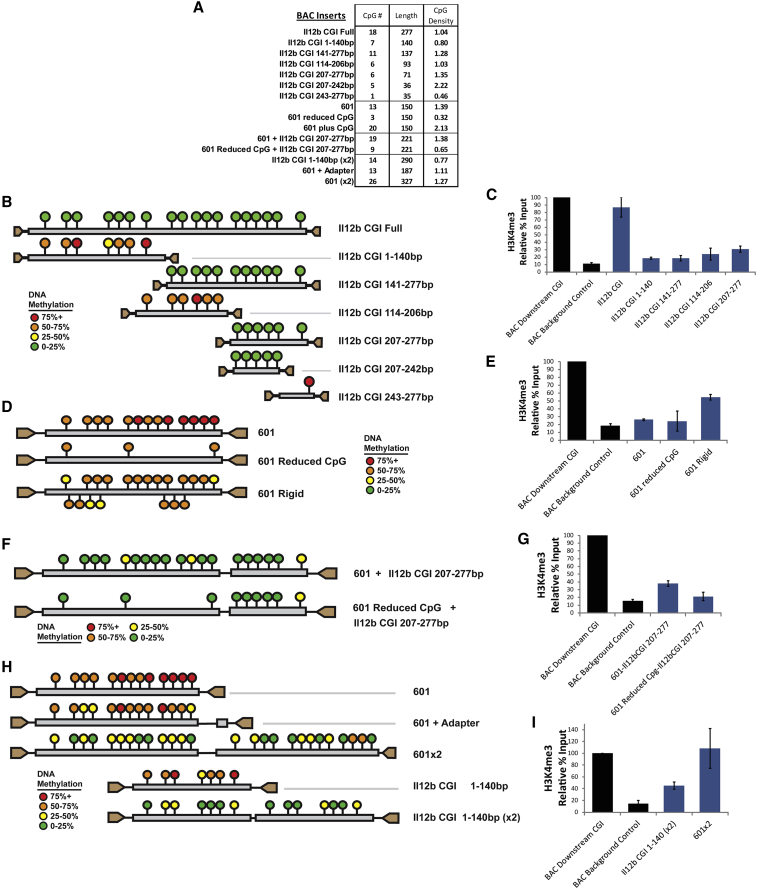
(A) The table displays the basic DNA properties of CpG-rich sequences inserted into a gene-desert BAC, which was then pre-methylated and introduced into mouse ESCs.
(B) The diagrams represent the DNA methylation status in ESCs at inserts of the Il12b CpG island and deletion variants. Each circle represents a particular CpG position, the color of which represents the average ratio of methylated to unmethylated CpGs determined by bisulfite sequencing across multiple clones. The data shown for each insert are an aggregate of three or four clones. The key shows the color that corresponds to each DNA methylation level.
(C) The graph shows qPCR results from H3K4me3 ChIP DNA with primers specific to select inserts from (B). The signal is relative to the input fraction and normalized to a native unmethylated downstream CpG island on the transgenic BAC. The signal at a control region 2 kb upstream of the insertion site is also shown. The black bars represent the standard error from two immunoprecipitation replicates of at least two clones.
(D) The diagrams show the DNA methylation status at transgenic inserts of the 601 sequence and variants, similar to (B).
(E) The graph shows qPCR results from ChIP for H3K4me3 with primers specific to the 601, “601 reduced CpG,” and “601 plus CpG” variant inserts in ESCs. Normalization and controls are as in (C).
(F) The diagrams show DNA methylation levels determined by bisulfite sequencing at transgenic inserts of either 601 or “601 reduced CpG” fused to the Il12b CpG island 207–277 bp sequence from (B).
(G) The graph shows qPCR results from H3K4me3 ChIP DNA with primers specific to the inserts of 601 and “601 reduced CpG” fusion variant fusions to the Il12b CpG island 207–277 bp fragment.
(H) The diagrams show DNA methylation levels at transgenic inserts of tandem 601 (601 (×2)) or Il12b 1–140 bp fragments (Il12b 1–140 bp (×2)). Also shown are DNA methylation levels at an insert composed of 601 plus the adaptor used to adjoin tandem sequences.
(I) The graph shows qPCR results from H3K4me3 ChIP DNA with primers specific to the tandem insertions in (H).