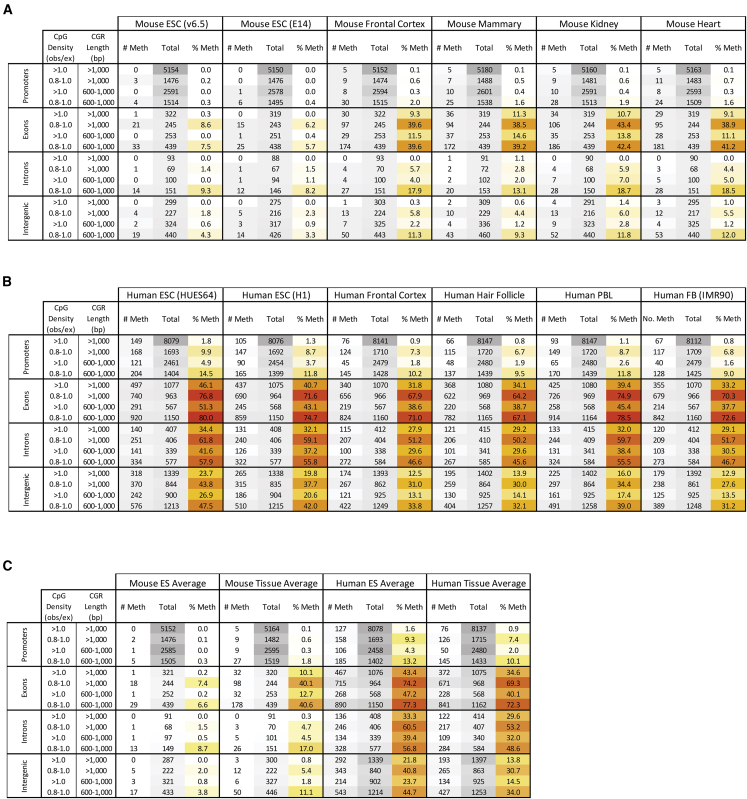
Figure 3.
Prevalence of Highly Methylated CGRs Identified Using Defined CpG Density/CGR Length Criteria in Mouse and Human ESC and Differentiated Cells
(A and B) The tables show the counts of CGRs in mouse and human DNA methylation datasets that meet the DNA property criteria labeled at left. CGRs are separated by genomic location and by non-overlapping CpG density and length ranges. Cell line names are in parentheses, with all other datasets derived from primary tissues. For all six mouse (A) and six human (B) datasets, the number of CGRs with high DNA methylation (>70%) in each criteria range is shown under “# Meth” next to the number of regions that qualify for the criteria, “Total.” The last column for each methylome group is the percentage of CGRs methylated for each criterion, “% Meth.” The tables are colored by frequency; increased gray indicates higher CGR numbers, while increased red indicates a higher percentage in the “% Meth” column. CGRs with insufficient bisulfite sequencing reads were discarded, which accounts for the slight variation in qualifying CGRs between methylomes. In addition, promoter CGR classifications were confirmed manually for all methylated mouse CGRs shown and all methylated human CGRs >1.0 CpG density and >1 kb CGR length (see Figure 4). PBL, peripheral blood leukocytes; FB, fibroblasts.
(C) Averaged and rounded values for the mouse and human ESC and somatic cell data from (A) and (B) are shown.