Figure 4.
The Relationships between DNA Properties and DNA Methylation Differ in Mice and Humans and during Development
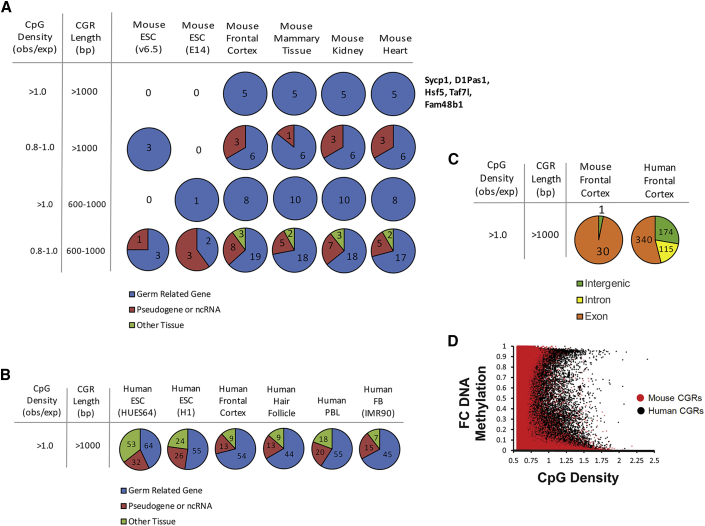
(A) The pie charts show the gene types associated with the highly methylated mouse promoter CGRs identified using the DNA property criteria at the left (see also Figure 3). Each graph shows the number of CGRs for each mouse cell type that have an associated gene that is either germline related, pseudogene/non-coding, or non-germ related (skin, neuronal, etc.). The five CGRs with CpG density >1.0 and length >1 kb and that have high methylation are from the same five genes in all four differentiated cell types; the names of these five genes are displayed to the right.
(B) The pie charts show the characterization of the confirmed promoter human CGRs with CpG density >1.0 and length >1 kb that are methylated from each cell type, as in (A).
(C) The pie charts show a comparison of the methylated mouse and human CGRs with CpG density >1.0 and length >1 kb, at non-promoter locations in frontal cortex. Each graph shows the number of methylated CGRs at exons, introns, or intergenic regions, colored by the key at the bottom.
(D) The dot plot shows the distribution of average DNA methylation in frontal cortex at all CGRs compared with CpG density, for both mouse (red) and human (black).