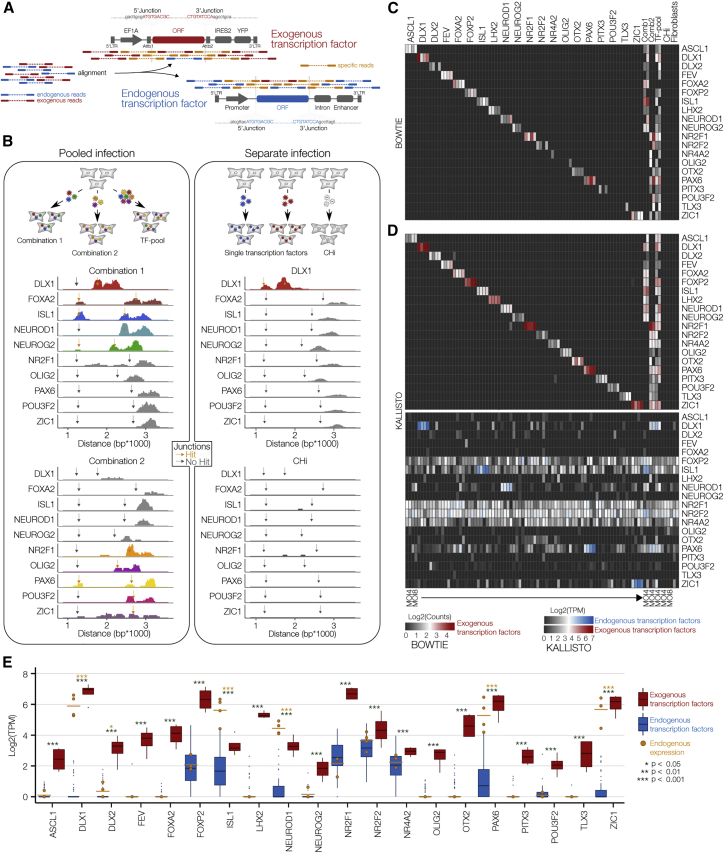
Figure 3.
Distinguished Detection of Exogenous and Endogenous Transcripts
(A) Schematic depicting the strategy to distinguish exogenous and endogenous sequencing reads.
(B) Bulk RNA-seq on pooled and separately infected fibroblasts. Horizontal dimension, distance from the 5ʹ end of the EF1A promoter; vertical dimension, number of aligned paired-end reads. Gray arrows (no overlap) and golden arrows (overlap) mark 5ʹ and 3′ junctions of exogenous ORFs.
(C and D) Heatmaps showing log2-transformed count values of exogenous TFs after alignment using Bowtie (C) and log2-transformed tags per million (TPM) values of exogenous and endogenous TF pairs after trimming junction sequences to ~100 base pairs and alignment using Kallisto (D). For individually infected fibroblasts and CHi, two replicates at an MOI of 4 and two replicates at an MOI of 8 were included. For pooled infected fibroblasts, two replicates at an MOI of 4 were included.
(E) Boxplots showing increased exogenous (red) versus endogenous (blue) expression of all TFs across all individually infected fibroblasts. Golden dots show endogenous expression in samples infected with the corresponding exogenous TFs. Unpaired Student's t test. Error bars represent mean + SD.