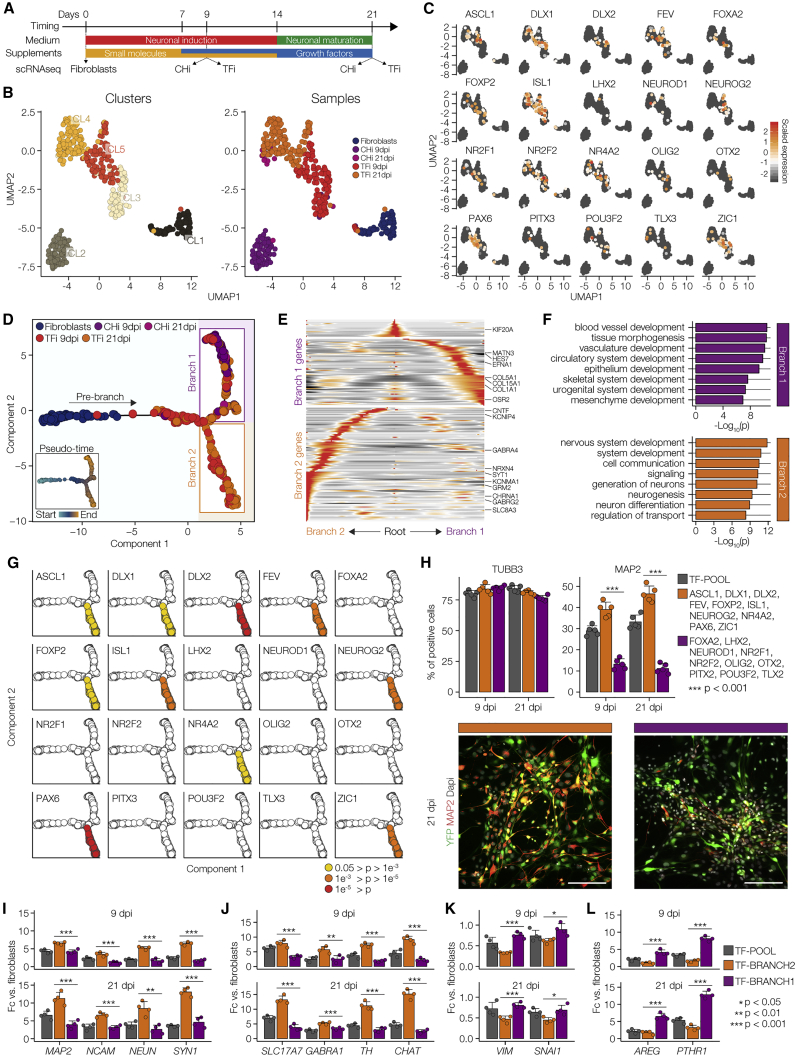
Figure 4.
Association of Developmental Trajectories with Exogenous Expression Profiles
(A) Diagram of the differentiation protocol of TFi and CHi, depicting the samples used for the time course Smart-seq experiment.
(B) UMAP 2D cell maps of the time course data. Left: cells were colored by cluster identity. Right: cells were colored by sample identity. Fibroblasts (n = 78 cells), CHi at 9 dpi (n = 87 cells), TFi at 9 dpi (n = 129), CHi at 21 dpi (n = 15 cells), and TFi at 21 dpi (n = 137 cells).
(C) Visualization of relative expression values of exogenous TFs on UMAP plots.
(D) Pseudo-temporal ordering of the Smart-seq time course based on the expression of 2,925 developmental genes (n = 446 cells). Small inset shows the same plot colored by pseudo-temporal values.
(E) Heatmap showing ~200 genes with branch-specific differential expression as determined by BEAM (R Package “Monocle”). In this heatmap, columns are points in pseudo-time, rows are genes, and the middle (Root) is the beginning of pseudo-time. Branch 1 goes from the middle of the heatmap to the right, while branch 2 goes to the left.
(F) GO analysis of genes differentially expressed in branch 1 (top panel) and branch 2 (bottom panel).
(G) Identification of exogenous TFs with branch-specific enrichment based on Fisher's exact tests.
(H) Top panels: quantification of TUBB3+ and MAP2+ cells in fibroblasts infected with the complete TF-pool (gray), branch 2-enriched TFs (orange), and unenriched TFs (purple) at 9 and 21 dpi. n = 5 independent experiments, unpaired Student's t test. Error bars represent mean + SD. Bottom panels: representative images of immunostainings for MAP2 (red) at 21 dpi; colors as in top panels. YFP (green) marks infected cells and cell nuclei were visualized using DAPI nuclear stain (gray). Scale bars, 100 μm.
(I–L) Neuronal differentiation, loss of fibroblast characteristics, and acquisition of alternative developmental fates as revealed by qPCR for pan-neuronal marker genes (MAP2, NRCAM, NEUN, SYN1) (I), canonical neuronal subtype markers (VGlut1, GABA, TH, CHAT) (J), canonical fibroblast markers (VIM, SNAI2) (K), and branch 1-enriched genes (AREG, PTHR1) (L) at 9 and 21 dpi; colors as in (H). n = 4 independent experiments, unpaired Student's t test. Error bars represent mean +SD.