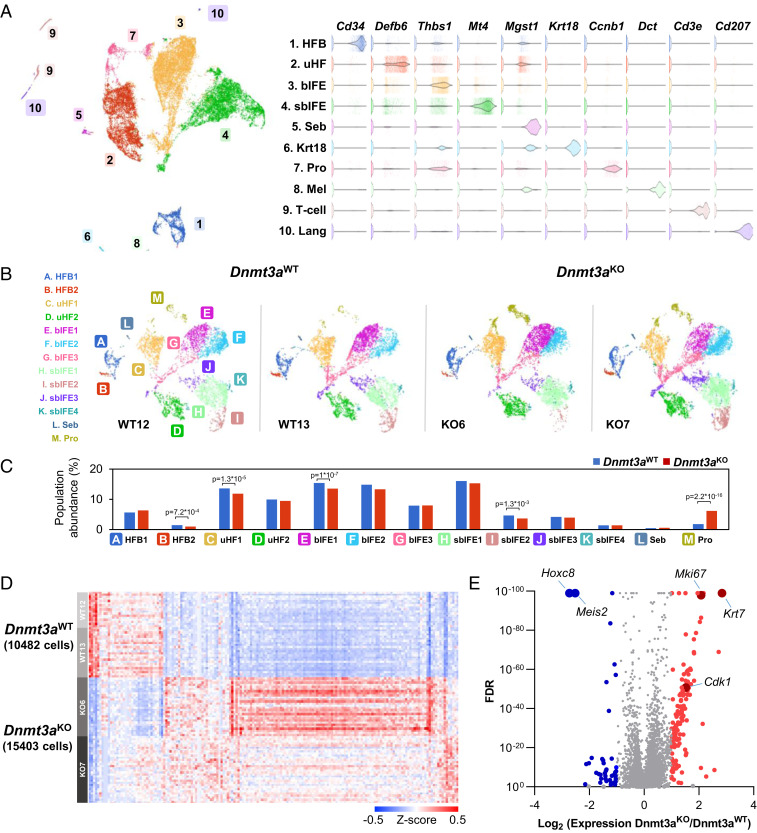
Fig. 2.
Single-cell RNA sequencing reveals that Dnmt3a deficiency causes discrete gene expression changes in murine keratinocytes. (A) UMAP representation of single-cell RNA sequencing data from whole epidermis of two pairs of Krt14-Cre− × Dnmt3afl/fl (Dnmt3aWT; 10,783 cells) and Krt14-Cre+ × Dnmt3afl/fl mice (Dnmt3aKO;16,324 cells) with unbiased graph-based clustering demonstrating known skin populations represented by lineage defining genes (Right), including HFB, uHF, bIFE, sbIFE, and sebaceous gland (Seb), Krt18-expressing cells (Krt18), proliferative cells (Pro), melanocytes (Mel), T cells (T-cell), and Langerhan’s cells (Lang). (B) Comparison of keratinocyte lineage cells in the Dnmt3aWT versus Dnmt3aKO mice, excluding inflammatory cells and melanocytes (excluded clusters 6, 8, 9, and 10). UMAP projections for each pair of samples with the indicated genotypes are shown. (C) No major population shifts are observed in epidermal lineages affected by Krt14-Cre mediated Dnmt3a deficiency, with the exception that the cells dominated by a proliferative signature (Pro, cluster M) are increased (6.1% in Dnmt3aKO versus 1.8% in Dnmt3aWT), as indicated by the bar graphs of keratinocyte lineage subclusters. Subclusters of the hair follicle bulge (A and B), upper hair follicle (C and D), basal interfollicular epidermis (E–G), suprabasal interfollicular epidermis (H–K), and sebocytes (L) are relatively unchanged. (D) Canonical gene expression changes were noted in single-cell RNA sequencing comparing keratinocyte lineage cells in Dnmt3aWT versus Dnmt3aKO epidermis (n = 2 mice per genotype). (E) There are 174 differentially expressed genes between Dnmt3aWT and Dnmt3aKO keratinocyte lineage cells with 137 up-regulated in the Dnmt3aKO and 37 down-regulated genes with fold change >2 and FDR <0.01.