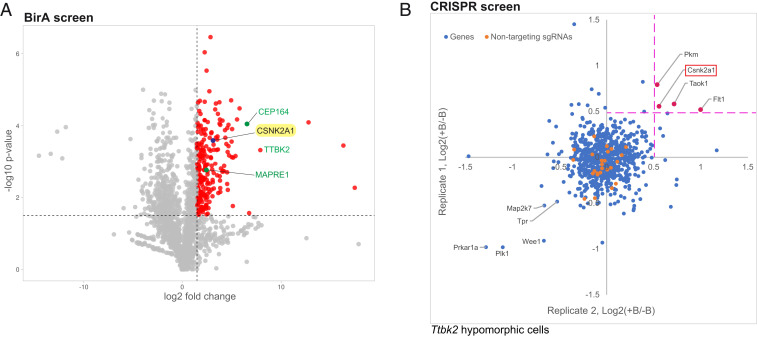
Fig. 1.
Identification of CSNK2A1 as a putative TTBK2-interacting protein. (A) Volcano plot of TTBK2-proximate proteins labeled by BioID. CEP164, TTBK2, and MAPRE1, known interactors of TTBK2, are shown in green. CSNK2A1, a candidate TTBK2-proximate protein is highlighted in yellow. The x-axis denotes the log2 fold change of TTBK2:GFP control. In the y-axis, the significance displays the negative log10-transformed P value for each protein. The red dots show the proteins considered as hits, delimited by the selected thresholds ≥ 1.5 for the log2 fold change and statistical significance. (B) The CRISPR screen plot for Ttbk2null/gt cells. The suppressor screen identifies potential negative modifiers of TTBK2, including CSNK2A1 (red). Each gene is plotted using the mean of medians values (defined as the two median ratios of sgRNAs counts, calculated as follows: +blasticidin(+SAG): −Blasticidin(+SAG); note that four sgRNAs represent each gene). Orange dots represent the nontargeting sgRNAs.