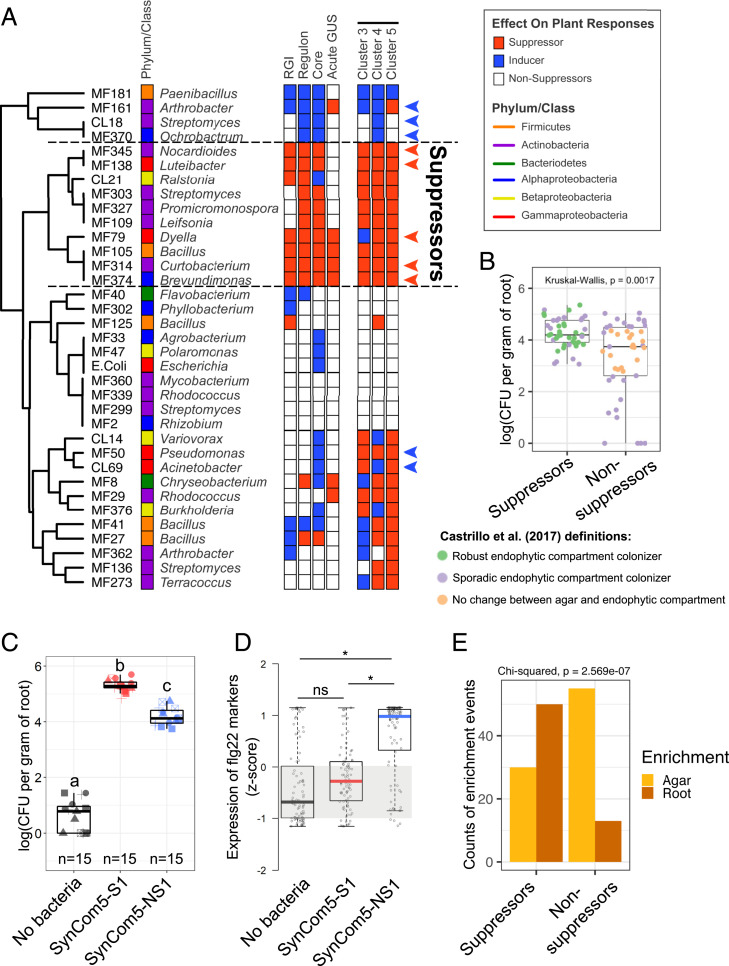
Fig. 3.
Microbiota members that suppress the plant immune system are better colonizers of Arabidopsis roots either in monoassociation or in the context of SynComs. (A) Summary of the assays performed to define suppression of the flg22 response in this study. Strains are clustered based on their response to a total of five assays that are illustrated as rows of boxes to the right. RGI: the ability of a strain to suppress flg22-induced RGI or induce this phenotype even in the absence of the MAMP (Fig. 2A). Regulon: the ability to interfere with the overall expression signature of 428 flg22-responsive genes defined in control (no bacteria) seedlings (Fig. 2B). Core: the ability of each strain to interfere specifically with a set of 84 defense-related genes that are consistently induced by flg22 under diverse experimental conditions (SI Appendix, Fig. S6C). Acute GUS: the ability of each strain to interfere with the acute response to flg22 (5 h treatment) specifically in roots based on the pCYP71A12::GUS reporter line (SI Appendix, Fig. S6D). Clusters 3, 4, and 5: the suppression ability of each strain on specific transcriptional response sectors of the flg22 regulon (Fig. 2C). Members of SynCom5-S1 and SynCom5-NS1 are marked with red and blue arrowheads, respectively. (B) Suppressors are better colonizers of Arabidopsis roots in monoassociation experiments. The growth of five suppressors (members of SynCom5-S1) and taxonomically matching nonsuppressors (members of SynCom5-NS1) were individually assayed in Arabidopsis roots after 12 d of colonization in three biological replicates. Each dot represents an individual strain/replicate. Strains are colored based on their behavior in the context of SynCom35 in a previous study (22). Note that all robust endophytic compartment colonizers are suppressor strains. A Kruskal–Wallis test was performed on the CFU counts from three independent experiments each with three technical reps. n = 9 for each strain and n = 45 for each group being compared. (C) A SynCom made of five suppressors (SynCom5-S1) colonizes Arabidopsis roots better than a SynCom made of five taxonomically matching nonsuppressors (SynCom5-NS1). Different symbols represent independent experimental repetitions. Letters indicate significantly different groups based on an ANOVA followed by a Tukey test (α = 0.05). (D) Seedlings grown with SynCom5-NS1 activate the expression of flg22 marker genes while seedlings inoculated with SynCom5-S1 do not. Statistical significance was determined using a permutation approach, where the actual group mean differences were compared to the group mean differences of 10,000 permutations of the gene counts. The group mean differences that were greater than 95% of the group mean differences calculated in the permutations were considered significant (asterisks). n.s. = not significant. (E) Suppressor (SynCom5-S1), but not nonsuppressor (SynCom5-NS1), strains are enriched in the endophytic compartment of Arabidopsis roots relative to agar in the context of a synthetic community. Suppressors n = 80, nonsuppressors n = 68.