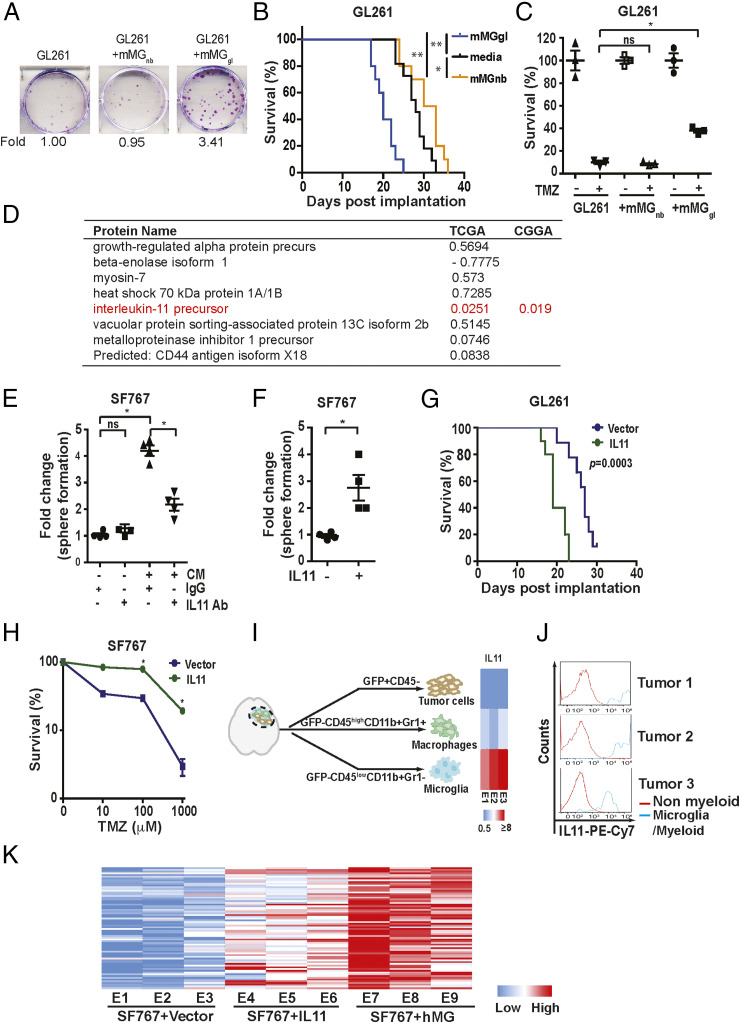
Fig. 2.
Glioblastoma-associated microglia release IL11 to enhance glioblastoma tumorigenicity and TMZ resistance. (A, Upper) Representative images of in vitro colony formation after coculturing of freshly isolated mMGgl or mMGnb with murine GL261 glioblastoma cells. (A, Bottom) Fold change in colony-forming units with or without mMGgl or mMGnb coculturing. (B) Effect of orthotopic coimplantation of mMGgl or mMGnb and GL261 on mice survival. *P < 0.05; **P < 0.0001. n = 10. (C) The effects of coculturing of GL261 with mMGgl or mMGnb on TMZ resistance, assessed by colony formation assays. *P < 0.05. (D) The 10 most abundant proteins identified through unbiased proteomic profiling of CM derived from immortalized hMG. P value of association between mRNA expression of genes encoding these CM proteins and survival in the TCGA are shown under the TCGA column. To avoid multiple comparisons, only IL11 was tested for survival association using the CGGA. P value for this association is shown under the CGGA column. (E) Effect of IL11 neutralizing antibodies on hMG CM-induced neurosphere formation in SF767. *P < 0.05. ns: not significant. (F) The effect of recombinant human IL11 (20 ng/mL) on the neurosphere-forming capacity of SF767 cells. *P < 0.05. (G) Survival of mice bearing GL261 cells ectopically expressing IL11 compared to mice bearing GL261 cells. n = 10. P = 0.0003. (H) Effect of ectopic IL11 expression on TMZ resistance of SF767 cells. In vitro cell viability was determined. *P < 0.05. n = 6. Error bars, SD. (I) GFP-labeled GL261intracranial tumors were sorted into tumor cells (GFP+CD45−), TAM (GFP-CD45highCD11b+Gr1+), and microglia (GFP-CD45lowCD11b+Gr1−). IL11 levels were examined by qRT-PCR, and the results were displayed as a heatmap. (J) IL11 levels in nonmyeloid cells (IBA1−CD68−CD11b−) and tumor-associated microglia/myeloid cells (IBA1+CD68+CD11b+) sorted from three glioblastoma specimens resected from human subjects. (K) The effect of microglia coimplantation and ectopic IL11 expression on the expression of the 82 survival-associated genes. RNA was extracted from subcutaneous tumors formed by SF767-vector, SF767-IL11, and SF767 cells coimplanted with microglia, and gene expression was analyzed by qRT-PCR. ΔCt between gene and actin in each sample was plotted as a heatmap. Three independent tumors were analyzed for each cohort.