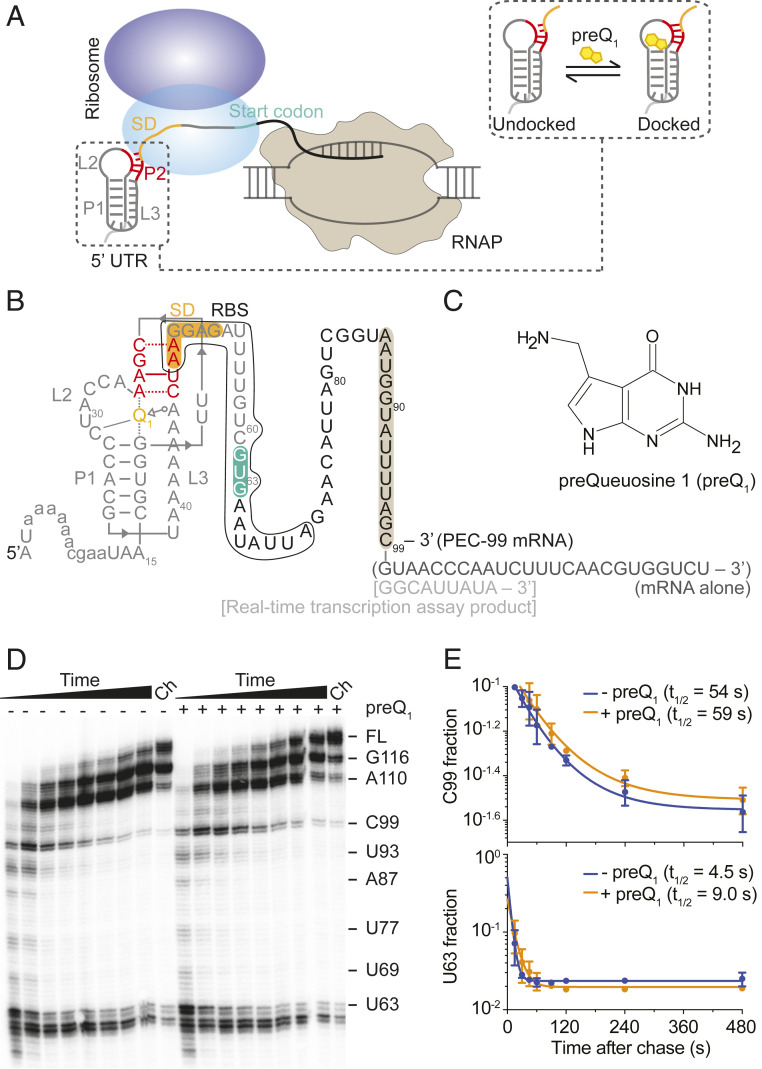
Fig. 1.
Bas mRNA containing a preQ1 riboswitch in the 5′ UTR as a model to study transcription–translation coordination. (A) Schematic representation of transcription–translation coupling on a nascent mRNA containing a structured riboswitch in the 5′ UTR. Binding of the cognate ligand to the riboswitch stabilizes a folded conformation where the SD sequence becomes less accessible to ribosome binding (shown in a dashed box). (B) Secondary structure of the Bas mRNA in our system showing the riboswitch pseudoknot at the 5′ end, the entire RBS, SD sequence (orange), and start codon (green) as indicated. Annotations in parentheses and square brackets indicate modifications introduced for experiments with mRNA alone and during real-time transcription assays, respectively. Sequence protected by the RNAP is highlighted in brown. (C) Chemical structure of the preQ1 ligand. (D) In vitro transcription of Bas mRNA. U63, C99, and full-length RNAs are shown on the right. The Ch lane corresponds to the final chase where 500 µM rNTPs were added at the end and the reaction was incubated for 5 min. (E) Quantification of the half-lives of the C99 and U63 pause sites in the absence or presence of preQ1.