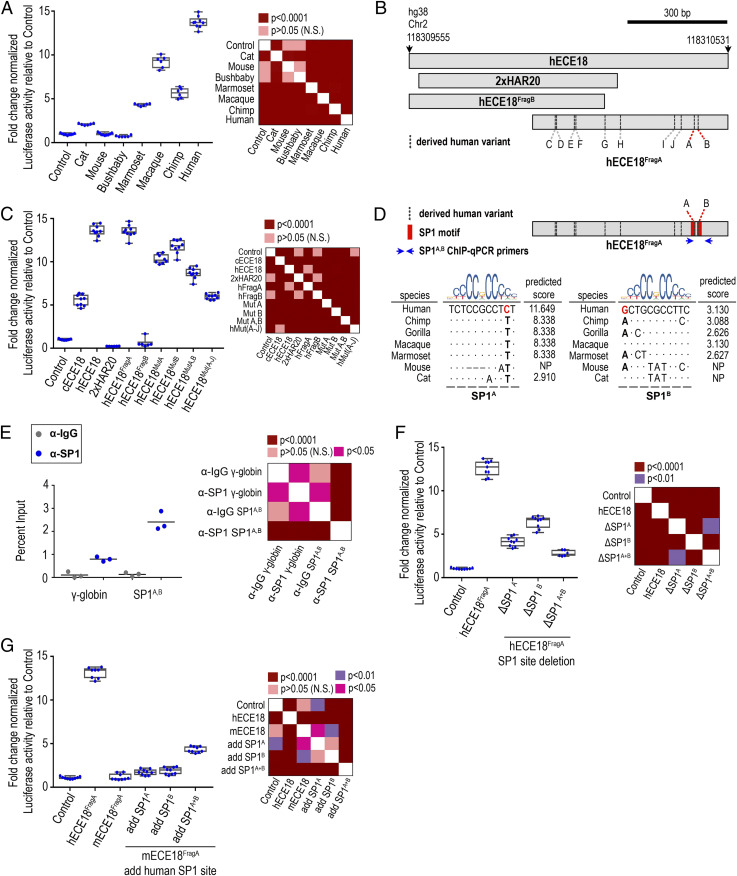
Fig. 2.
Repeated mutation of ECE18 produced human-specific gains in enhancer activity. (A) Fold induction of luciferase reporter activity relative to empty vector (Control) by ECE18 orthologs in cultured human keratinocytes. (B) Schematic of ECE18 variants tested in C. Derived human bases at A and B are highlighted in red, and the positions of the remaining derived human-specific base substitutions are shown as dashed vertical lines. (C) Localization of hECE18 enhancer activity. The fold change normalized luciferase activities of full-length hECE18, chimpanzee ECE18 (cECE18), human 2xHAR20, hECE18 fragment (Frag) A and B, and mutant hECE18 in which the indicated derived base is replaced by the ancestral ape base are plotted. Mutated base (Mut). All 10 derived human substitutions are mutated to ancestral ape bases in hECE18MutA−J. (D) Species alignment and predicted binding affinities for SP1 at SP1A and SP1B. Derived human bases at A and B are in red. Dots indicate identity to human base. Blue arrows indicate the location of ChIP-qPCR primers used in E. (E) Enrichment of SP1 by ChIP-qPCR at hECE18 interval containing SP1A and SP1B motifs (SP1A, B) in human keratinocytes. Human γ-globin (HBG2) promoter is used as a negative control, and IgG or SP1 enrichment over the input for each set of primers is shown. Mean enrichment is reported across three independent experiments (line). (F) Fold induction of luciferase reporter by hECE18FragA upon deletion of SP1A and SP1B. (G) Fold induction of luciferase reporter by mouse ECE18FragA (mECE18FragA) in human keratinocytes upon knock-in of human SP1A and SP1B motifs. Normalized Firefly luciferase activity is plotted as the fold change relative to Control (empty reporter vector). Firefly luciferase values are normalized to Renilla luminescence. The dots represent an individual biological replicate. Median (line), box (bounds 25 to 75%) and whiskers (min and max) plotted. Significance by one-way ANOVA. Tukey-adjusted P values are reported in heatmaps. Assays are performed in human GMA24F1A keratinocytes.