FIGURE 4.
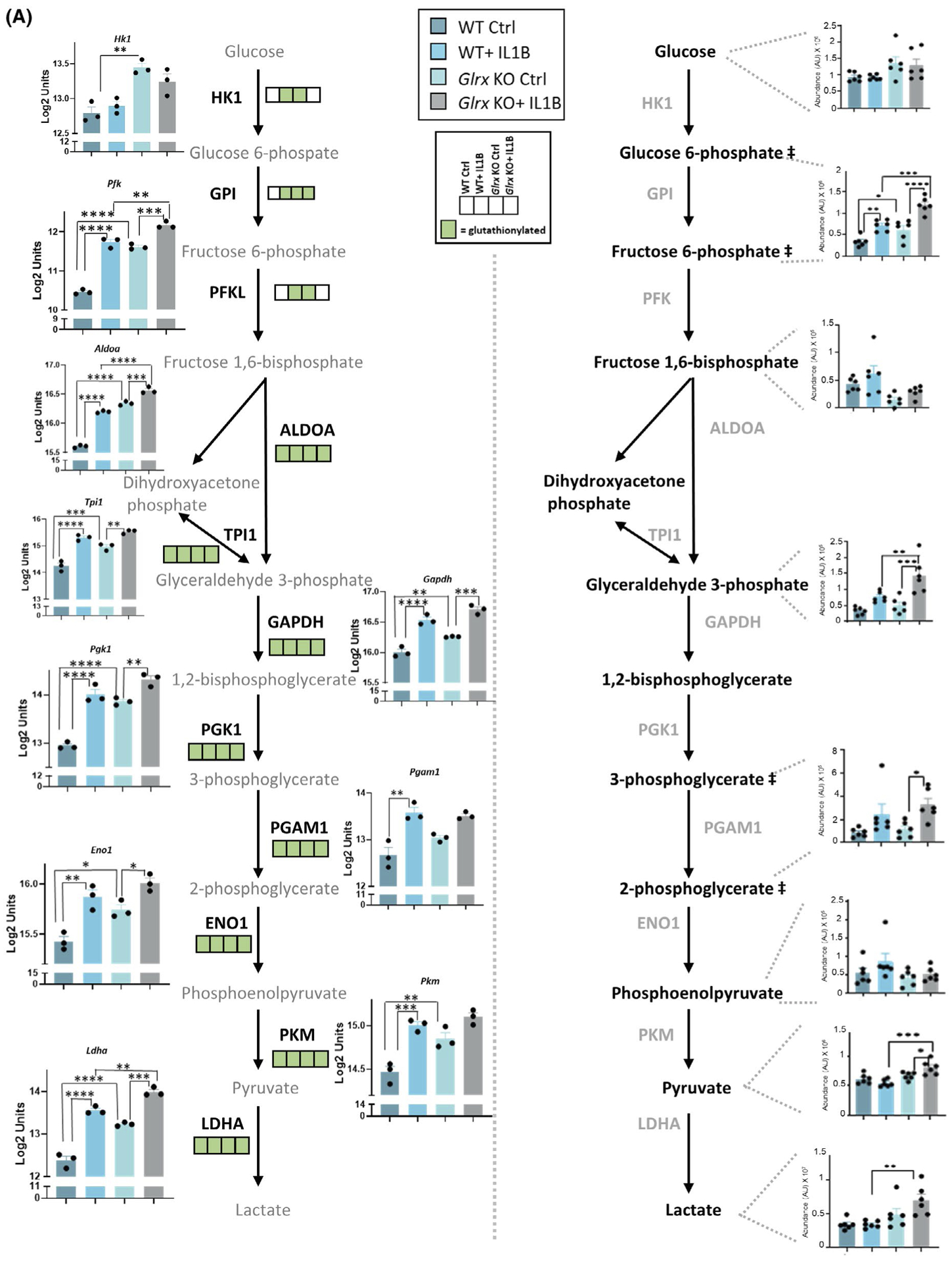
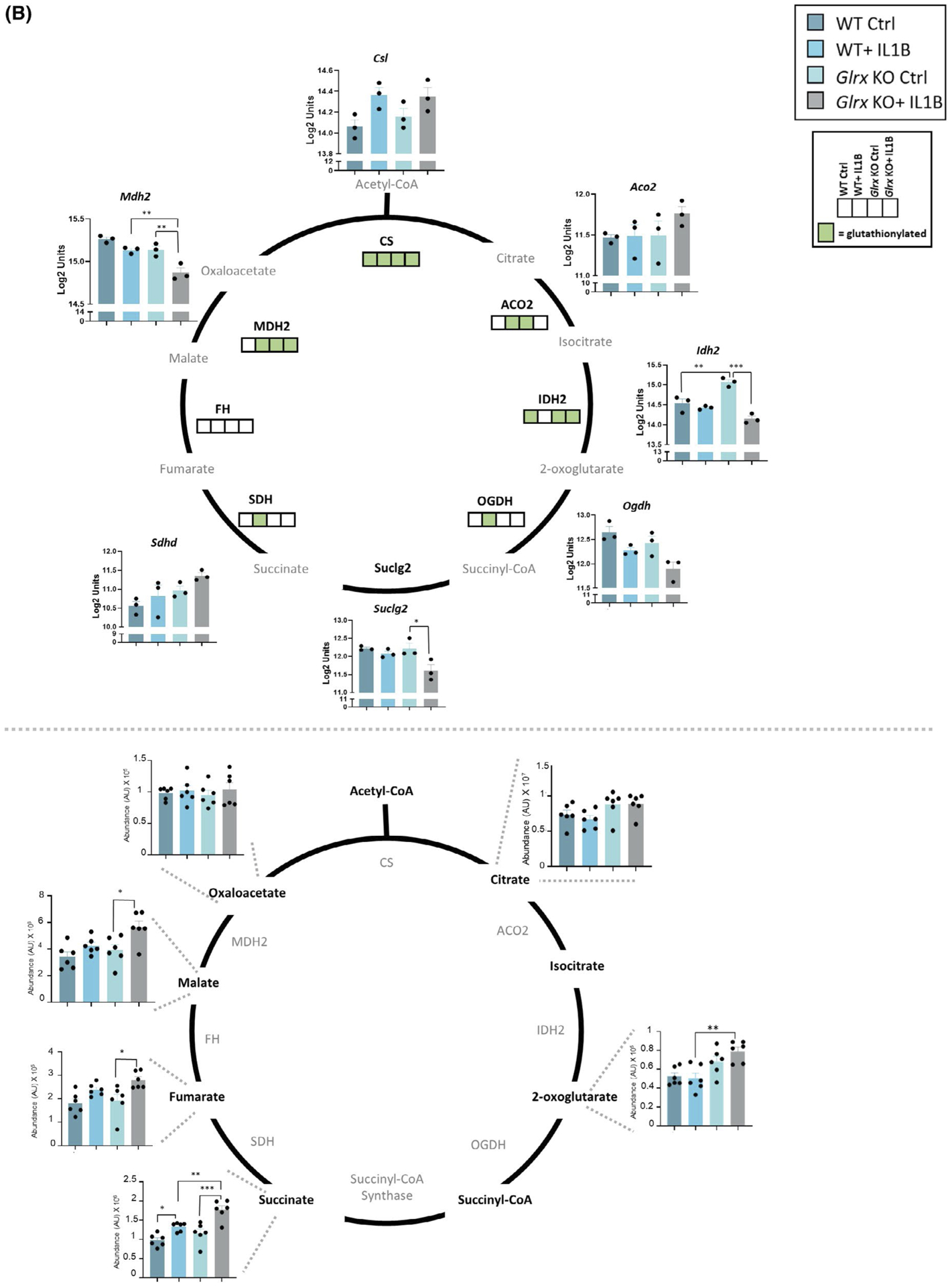
Multi-omics integration of KEGG pathways affected in airway basal cells stimulated with IL1B that are affected by GLRX status. Visualization of hits in the TCA cycle and glycolysis pathway identified in the -omics platforms described in Figures 1–3. A, Glycolysis pathway; right panel shows the total metabolites and left panel shows the microarray gene expression data as well as glutathionylated protein hits in each of the four groups (present; green, absent; white). ## Metabolites measured are hexose-6 phosphate (top) and phosphoglycerate (bottom). Hk1: hexokinase1, Pfk: phosphofructokinase, Aldoa: fructose-bisphosphate aldolase A, Tpi1: triosephosphate isomerase, Pgk1: phosphoglycerate kinase1, Gapdh: glyceraldehyde-3-phosphate dehydrogenase, Pgam1: phosphoglycerate mutase1, Eno1: enolase1, Pkm: pyruvate kinase M, Ldha: lactate dehydrogenase A, HK1: hexokinase1, GPI: glucose-6-phosphate isomerase, PFK: phosphofructokinase, ALDOA: Fructose-bisphosphate aldolase, TPI1: triosephosphate isomerase, GAPDH: glyceraldehyde-3-phosphate dehydrogenase, PGK1: phosphoglycerate kinase1, PGAM1: phosphoglycerate mutase 1, ENO1: alpha-enolase, PKM: pyruvate kinase M, LDHA: L-lactate dehydrogenase A B, TCA cycle; bottom panel shows the total metabolites and top panel shows the microarray gene expression data as well as glutathionylated protein hits in each of the four groups (present; green, absent; white). Csl: citrate synthase, Aco2: aconitate hydratase, mitochondrial, Idh2: isocitrate dehydrogenase, mitochondrial, Ogdh: 2-oxoglutarate dehydrogenase, mitochondrial, Suclg2: succinate coA ligase, subunit G, Sdhd: succinate dehydrogenase, mitochondrial, Mdh2: malate dehydrogenase, mitochondrial. CS: citrate synthase, ACO2: aconitate hydratase, IDH2: isocitrate dehydrogenase, OGDH: 2-oxoglutarate dehydrogenase, SDH: succinate dehydrogenase, FH: fumarate hydratase, MDH2: malate dehydrogenase, mitochondrial. *P < .05, ** P < .01, *** P < .001, **** P < .0001 (ANOVA)
