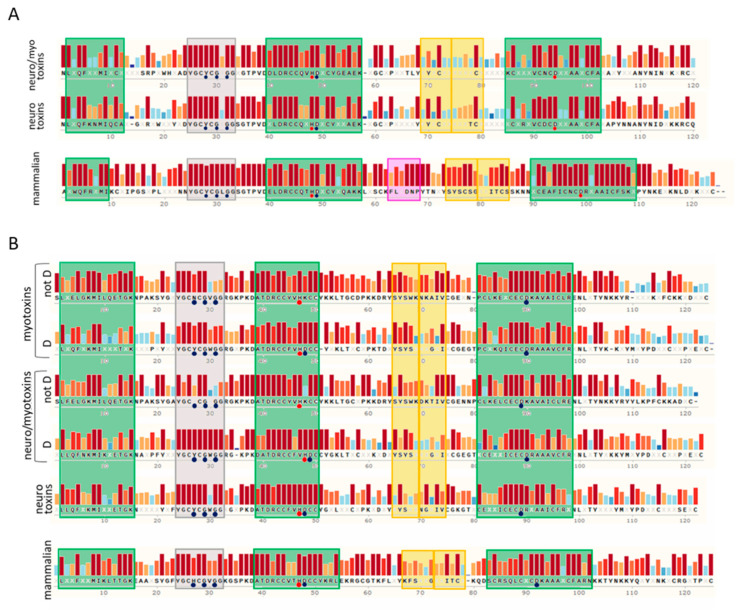
Figure 2.
Consensus sequences analyses of snake venom PLA2s and their mammalian homologs. (A) Consensus sequences of group I snake venom neuro-myotoxic and neurotoxic PLA2s compared with that of mammalian PLA2G1B. (B) Consensus sequences of group II snake venom myotoxic, neuro-myotoxic, and neurotoxic PLA2s compared with that of mammalian PLA2G2A. In the scheme, myotoxins and neuro-myotoxins are divided in the “not D” group and “D” group, according to the absence or presence of the D amino acid in the position known as “49” of the active site, respectively. The sequences were collected from the Swiss-Prot database and aligned with the align tool of UniProt (Clustal O, https://www.uniprot.org/align/, accessed on 1 March 2021). The alignments were then visualized with SnapGene Viewer version 4.3.11. Conserved residues are reported in the consensus sequence, whereas the X code indicates an amino acid. The colored bars above the protein sequence show the degree of conservation, where a red bar indicates the maximum level of conservation (100%). Green, yellow, and grey squares represent α-helix, β-sheet, and calcium binding loop, respectively. The pancreatic loop (pink square, panel A) is present only in mammalian PLA2G1B. Blue circles indicate amino acids involved in the Ca2+ binding, whereas red circles represent the catalytic sites. The alignments are reported in the Supplementary Material.