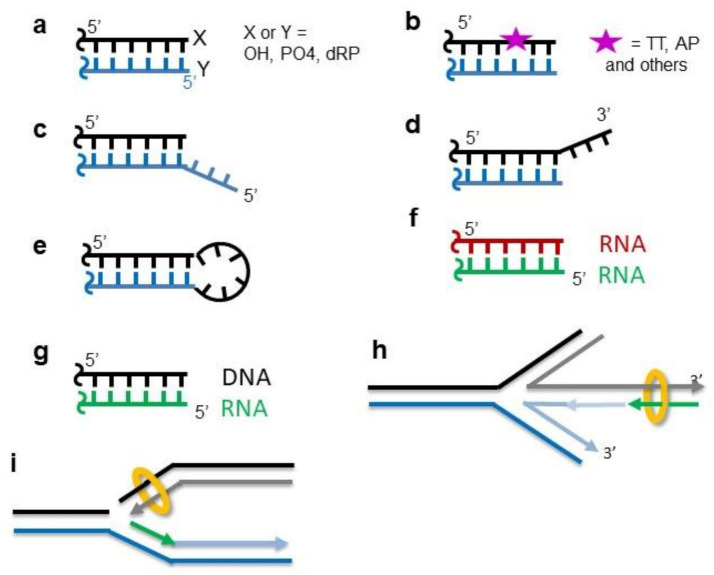
Figure 1.
Diversity of DNA ends that are recognized by Ku70/80. (a) Ku binds DNA ends efficiently with strands containing either hydroxyl or phosphate functions, as well as more heterogeneous ends with dRP or other chemical adducts. (b) Ku recognizes DSBs with a lesion present in the internal position close to the DNA ends. (c,d) Ku interacts efficiently with DNA having single-strand overhangs in 3′ and/or in 5′ up to 20 nt. (e) Ku can accommodate the presence of loops up to at least 8 nt at the ends. (f) Ku binds RNA hairpins at telomeres. (g,h) Ku interacts with the DNA–RNA duplex during replication fork restart (nascent RNA (green arrow)). (i) Ku binds single-ended DSB.