Figure 4.
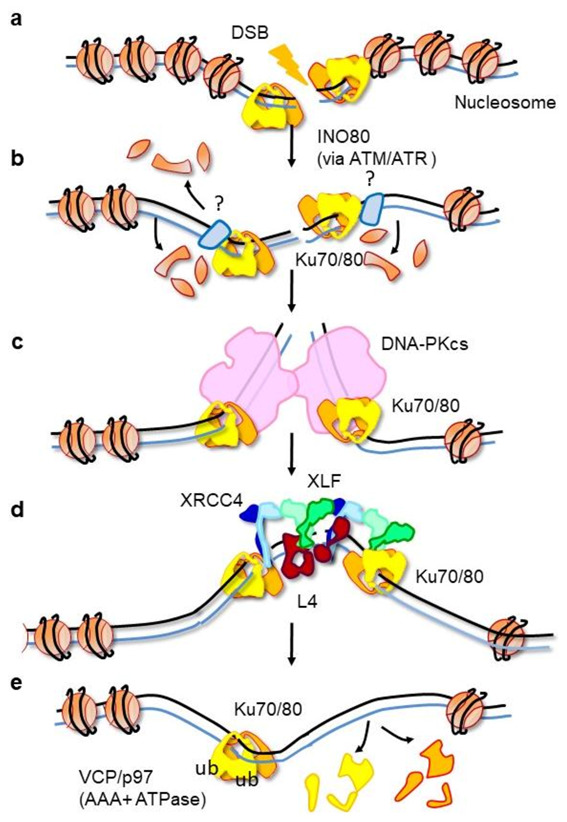
Association and removal of Ku in a chromatin context. (a) Upon DSB formation, Ku recognizes DNA ends. Super-resolution microscopy studies suggest that only one Ku may bind at the DSB and that no threading inward is observed with the loading of additional Ku molecules [52]. (b) Upon DSB formation, some nucleosomes are disassembled by the nucleosome remodeler INO80 [75]. Some factors not yet identified likely limit the Ku threading inward (light blue protein). c-NHEJ factors like XLF, APLF, or PAXX may be involved in this role. (c) Ku recruits DNA-PKcs, and the complex forms a synapse between the two DNA ends in a conformation that may correspond to the dimer observed by Chaplin et al. [11]. (d) Ku can recruit alternatively nucleases, polymerases, and the ligation complex at the DSB ends (here, the Ligase 4 is represented in a complex with an XLF-XRCC4 filament [104,105]. (e) Once the ligation is completed, Ku is trapped on the DNA. The AAA+ ATPase VCP/p97 will remove in place the trapped Ku molecules [101].
