Abstract
The world is currently facing a novel coronavirus (SARS-CoV-2) pandemic. The greatest threat that is disrupting the normal functioning of society is the exceptionally high species independent transmission. Drug repurposing is understood to be the best strategy to immediately deploy well-characterized agents against new pathogens. Several repurposable drugs are already in evaluation for determining suitability to treat COVID-19. One such promising compound includes heparin, which is widely used in reducing thrombotic events associated with COVID-19 induced pathology. As part of identifying target-specific antiviral compounds among FDA and world-approved libraries using high-throughput virtual screening (HTVS), we previously evaluated top hits for anti-SARS-CoV-2 activity. Here, we report results of highly efficacious viral entry blocking properties of heparin (IC50 = 12.3 nM) in the complete virus assay, and further, propose ways to use it as a potential transmission blocker. Exploring further, our in-silico analysis indicated that the heparin interacts with post-translational glycoconjugates present on spike proteins. The patterns of accessible spike-glycoconjugates in open and closed states are completely contrasted by one another. Heparin-binding to the open conformation of spike structurally supports the state and may aid ACE2 binding as reported with cell surface-bound heparan sulfate. We also studied spike protein mutant variants' heparin interactions for possible resistance. Based on available data and optimal absorption properties by the skin, heparin could potentially be used to block SARS-CoV-2 transmission. Studies should be designed to exploit its nanomolar antiviral activity to formulate heparin as topical or inhalation-based formulations, particularly on exposed areas and sites of primary viremia e.g. ACE2 rich epithelia of the eye (conjunctiva/lids), nasal cavity, and mouth.
Abbreviations: PBS, phosphate-buffered saline; FBS, fetal bovine serum; P/S, pen strep; ACE2, angiotensin-converting enzyme 2; NRP1, neuropilin-1; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; EMEM, Eagle's Minimum Essential Medium; CDC, centers for disease control and prevention; United States, FDA, U.S. Food and Drug Administration; PDB, protein data bank; RBD, receptor binding domain; TMPRSS2, transmembrane serine protease 2; UFH, unfractionated heparin; LMWH, low-molecular-weight heparin; HTVS, high throughput virtual screening; ID, identity
Keywords: COVID-19, SARS-CoV2, Heparin, Anticoagulant, Sulfated polysaccharide, Spike protein, Transmission-blocking, Lipid-based formulations, Topical delivery
1. Introduction
Heparin is one of the most administered medications in the world. It is part of the World Health Organization's (WHO) list of essential medicines [1,2]. Also known as unfractionated heparin (UFH), it is used as an anticoagulant, commonly used to treat heart attacks and unstable angina, and as a preventive measure for stroke. A fractionated version of heparin is also in common use, known as low molecular geruweight heparin. Since the beginning of the pandemic, clinical reports showed unique signs of coagulopathy associated with COVID-19 [[3], [4], [5], [6], [7], [8], [9], [10]], immediately prompting the need for medical professionals to consider tentative treatment. Unfractionated heparin (UFH) and low-molecular-weight heparin (LMWH) are widely used in the management of COVID-19 patients [7,[11], [12], [13]]. SARS-CoV-2 triggers an immense multiple systemic coagulation and inflammatory response, though it differs from traditional accounts of an immune response that leads to disseminated intravascular coagulation (DIC) [8]. This is because COVID-19-associated coagulopathy does not impair the ability to clot [9]. There are several parameters of coagulation that must be assessed in understanding the patho-immunology of COVID-19 progression. Some of these include measurements of D-dimer (DD), prothrombin time (PT), activated partial thromboplastin time (aPTT), fibrinogen, and antithrombin, all of which can hint at the direction and severity of coagulopathy in the patient [10].
Searching for repurposable drugs with potential targets among the drugs that are being evaluated as a treatment against COVID-19, heparin emerged as an interesting compound to pursue. The use of heparin for treating severe COVID-19-associated coagulopathy and DIC is rapidly becoming a routine patient management strategy. Though the exact mechanism is not fully understood, apart from reducing coagulopathy-related symptoms, it is likely that heparin also plays a role in reducing hypoxia, reducing cytokine storm intensity, as well as some antiviral effects. An earlier report has suggested the replacement of oral anticoagulant therapy with heparin in patients hospitalized with COVID-19, except for those patients that have a mechanical heart valve [14]. More clinical observations supported these findings in which the anticoagulant and anti-inflammatory action of heparin may be a useful treatment of pathological thrombotic events [15]. In addition to these effects, more studies are now suggesting that heparin may have some antiviral activity, though there is a limited understanding of the precise mechanism. To this end, recent preliminary results of several independent studies have shown that heparin can inhibit invasion of SARS-CoV-2 in a dose-dependent manner from 6.25–200 μg/μL [[16], [17], [18], [19]]. Apart from its known ability to inhibit various proteases such as furin and cathepsin-L, it is logical to hypothesize that heparin may be able to target the cellular entrance of SARS-CoV-2 [18,19]. Another plausible mechanism could be related to the spike (S) protein's interaction with neuropilin receptor-1, which is also facilitated by heparin. This then further prompts inquiry at the possible treatment options including both heparin and neuropilin receptor-1 [20]. Another study has shown that SARS-CoV-2 is dependent not only on ACE2 but also requires heparan sulfate proteoglycans [21]. The S protein has been shown to interact with both heparan sulfate receptors and heparin [21,22]. As such, studies have shown that heparin was able to abrogate virus infection to a similar extent as ACE2-specific antibodies [18].
In the present study, through the application of in-silico based virtual screening of FDA and world approved drugs [23] and specific in-vitro validations, we found that UFH had a sub-nanomolar activity and a potential to block receptor-binding domain (RBD) of SARS-CoV-2, thus pointing to its immense potential as a transmission-blocking agent. Collectively, the current findings emphasize the possible beneficial use of heparin, both as a transmission-blocking agent and treatment for coagulopathy/DIC associated symptoms in severe COVID-19 patients. Therefore, UFH presents itself as a critical multifaceted therapeutic option for COVID-19.
2. Methods
2.1. Antiviral activity of heparin (UFH) in a SARS-CoV-2 live virus assay
Test compounds were diluted to 2 times the indicated concentration in infection media (EMEM +2% FBS+ 1% P/S) and added to the cells for 1 h [24]. An equal volume of pre-titrated SARS-COV-2/MT020880.1 was added to cells at a multiplicity of infection (MOI) of 0.4 or 0.02, depending on entry or spread assay respectively, effectively diluting compounds to indicated concentration. After incubation for 1 h at 37 °C, the virus/compound inoculum was flicked off and washed 1× with PBS. Compounds at indicated concentrations were added back to the cells for 24 (entry assay) or 48 (spread assay) hour incubation at 37 °C. Cells were then fixed with 10% buffered formalin and permed with 0.2% Triton X for 10 min before blocking (BioLegend, Cat# 420201) overnight at 4 °C. Cells were immunostained by sequential incubation with SARS-CoV-2 antibody (SinoBiological, Cat# 40143-R001) and AlexaFluor 488 goat anti-rabbit (Invitrogen, Cat# A-11008) diluted in blocking buffer. Cell nuclei were stained with DAPI and imaged using an Operetta (Perkin Elmer) high content imaging instrument and infected cells were determined using Harmony Software (Perkin Elmer).
2.1.1. Spread assay vs. entry assay
For the spread assay, the virus used was at an MOI of 0.02 instead of 0.4 used in the Entry assay, and the assay was incubated for 48 h instead of 24 h used in spread assay.
2.2. In-silico target identification analysis
Around 36 crystallographic structures of spike protein are available at the Protein Data Bank (PDB). For our studies, we used PDB-ID.: 7CAI with a resolution of 3.49 Å, in complex with the two H014 Fab neutralizing proteins [25]. The SARS-CoV-2 S-trimer had two RBDs in the open state and one in the closed state. The protein was prepared using the Protein Preparation Wizard tool included in Maestro (Schrodinger Suite 2020–4). Water molecules, co-factors of crystallization, and ligands were removed, missing atoms were added, side chains and loops were filled by Prime, and hydrogens were added with Epik module options provided in protein preparations wizard at physiological pH. This final structure of the protein was minimized with the OPSL-3e force field as implemented in Maestro with an implicit solvent (water). The final minimized structure was used for docking purposes.
2.3. Ligand preparation
The ligands used in this study were heparin molecules (Semi-rigid Solution Structure of Heparin; degree of polymerization {dp} = 24) PDB-ID:3IRJ [26]. Representing the average molecular size of UFH, all 15 representative heparin models in the PDB entry were used for docking. The ligands were prepared by the protein preparation wizard to remove hydrogen binding errors and Lewis structure errors.
2.4. Docking calculations
These were carried out utilizing the Glide module from Schrodinger Suite [27,28]. Multiple grids encompassing the closed and open state of the spike protein were used for docking to accommodate large heparin molecules. The analysis was carried out in the first stage using the Standard Precision (SP) mode followed by an Extra Precision (XP) model which performs an advanced scoring, which in turn, results in an enriched calculation that minimizes false positives. The equation used to calculate the binding energy in the XP mode: XP Glide Score = Ecoul + Evdw + Ebind + Epenalty where Ecoul and EvdW represent van der Waals and electrostatic terms, respectively. End and Epenalty refer to contributions that favor binding or penalization of interactions that influence the binding of a ligand. The top lowest energy docked complexes were analyzed for binding patterns.
2.5. Mutant spike variants and heparin interactions
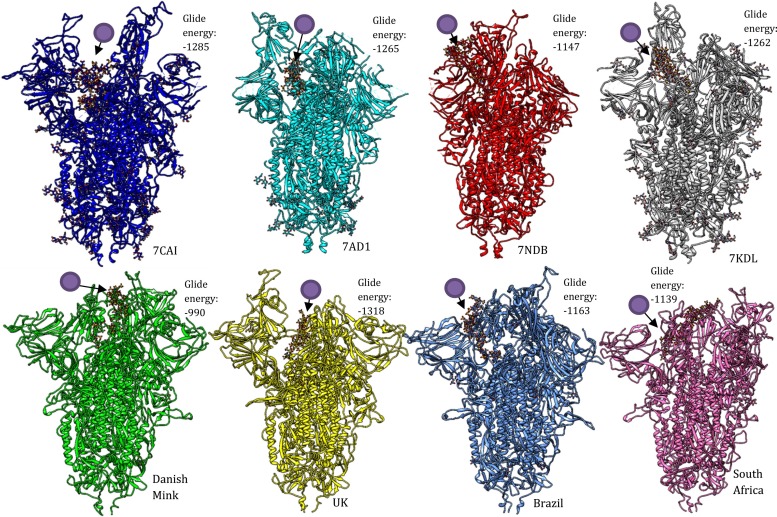
For our studies, we used PDB-ID.: 7AD1, 7NDB, and 7KDL representing different early mutants (Table 3). The regional variants supercomputer predicted 3D structures were downloaded from https://spikemutants.exscalate4cov.eu/#pdb which had different sets of polymorphisms including deletions (Table 3). The structures were prepared and docked with heparin molecules as described previously. Further mutation data was obtained from the GISAID database and epidemiological resource https://nextstrain.org/.
Table 3.
The reported mutants and their available structures. The region-specific variant's structures were predicted by supercomputers available from https://spikemutants.exscalate4cov.eu/#pdb funded by EU grant 101003551 [45].
| Common Name | Accession No./PDB ID | Lineage | Mutations | References |
|---|---|---|---|---|
| UK variant | VOC 202012/01 | B.1.1.7 lineage | HV 69–70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H. | [[46], [47], [48]] |
| South Africa variant | 501Y.V2 | B.1.351 lineage | L18F, D80A, D215G, R246I, K417N, E484K, N501Y, D614G and A701V. | [[49], [50], [51]] |
| Danish Mink variant | HV 69–70 deletion, Y453F, D614G, I692V, S1147L, and M1229I. | [52,53] | ||
| Brazilian variant | B.1.1.248 | B.1.1.28 | K417N/E484K/N501Y- L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, and V1176F. | [[54], [55], [56]] |
| Japan variant | GISAID ID: EPI_ISL_792680 to 792,683 | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, and V1176F | [57] | |
| D614G | 7KDL | D614G | [58] | |
| D614N | 7AD1 | D614N, R682S, R685G, A692P, A942P and V987P | [59] | |
| Engineered mutant (polybasic site) | 7NDB | K987P, V986P, R685S, R683S and R682G | [60] |
3. Results & discussion
Spike protein inhibitors have been investigated by many groups, and several vaccine candidates also depend on spike neutralization properties. Various heparin molecules have come up during virtual screening studies but are normally ignored due to their large size. A macromolecular agent like heparin would be ideal for blocking Spike-ACE2 interactions due to its known interactions with Spike and is inexpensive to produce, unlike antibodies. Tight interaction between these two molecules is due to the large size of the spike protein and the number of highly polymorphic glycoconjugates present on the protein surface (Fig. 3). In general, the experimental investigation of novel compounds is costly and requires an immense number of resources. However, the use of computational drug design applications, such as virtual screening, can be a time and cost-effective strategy to reduce the timeline and costs associated with the preclinical phase of drug development.
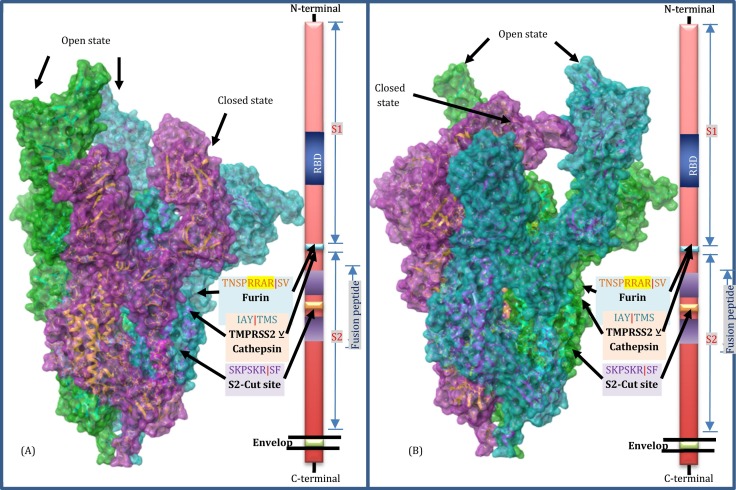
Fig. 3.
Schematic representation of spike trimer structure (PDB_ID:7CAI) [20]. Poses represented in Panel A and B are 180-degree rotation of the model on the vertical plane. Green and Turquoise surface representations are chains A and C with open state spike with RBD regions extended away from the main spike structure. The purple surface model of Chain B represents a closed state in which RBD is facing inwards and seems to be inactive for ACE2 binding. The processing sites are described according to earlier reports [74]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. In-silico drug discovery approach
In this study, a virtual screening through robust Glide module of Schrodinger software suite [29] was applied to predict spike binders preferring larger molecules binding to RBD-1 (ACE2 binding domain) or RBD-2 (NRP1 binding domain). We found two different binding poses with high binding energy against both closed (Glide energy: −183) and open states (Glide energy: −194) of the spike protein. The binding with the open state is in the trunk (S1-S2 junction) of the spike protein, on the opposite side of the RBD (Figs. 4A & 5F). This structural region (pose) appears to support the open state and possibly assist in ACE2 interactions. This could be the possible interaction responsible for cell-bound heparan sulfate, thus playing a crucial role in spike-ACE2 binding. On the other hand, in a closed state, heparin binds differently and the RBD domain is covered by heparin and the complex seems to be ‘locked in a closed state (Figs. 4B & 5A–E). This is in line with the previous reports on heparin sulfate being essential for spike binding to ACE2 [30] and blocking of SARS-CoV-2 by sulfated polysaccharides [18]. Moreover, the deletions and mutations now being found in various clinical SARS–CoV-2 isolates, lie in the S protein region near viral fusion domain 1 (S2), which is away from heparin binding to the closed state. It is important to note that single mutations are less likely to hamper macromolecular interactions with such a large interaction area. Interestingly, the fact that unfractionated heparin is like cellular heparan sulfate was recently reported to be critical for SARS-CoV-2 spike and ACE2 interactions [21,30]. Heparin being non-toxic (exception of rare complications and allergies) has been categorized as an essential therapeutic agent [2] and is being used as a topical application for several other ailments [31].
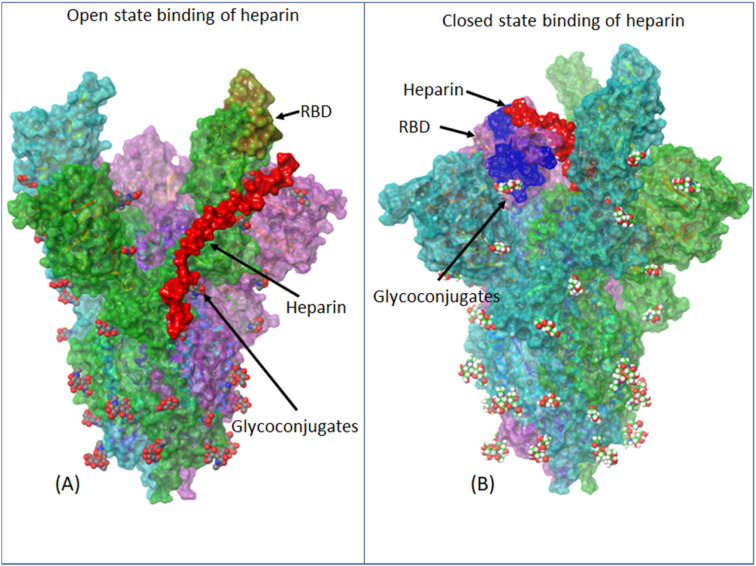
Fig. 4.
Molecular docking analysis of heparin against spike protein of SARS-CoV-2 envelope. The trimeric protein complex had two high scoring attachment sites (A) Heparin-binding to an open state i.e. RBD (brown highlights on green) is protruding exposing the ACE2 binding region. This binding is more supportive of spike-ACE2 interaction and possibly stabilizes the open state. This interaction could explain the role of heparan sulfate in facilitating ACE2 receptor binding as reported earlier [15] (B) heparin-binding to the closed domain: This binding has a similarity to open state binding as heparin molecule traces the sugar moieties (glycoconjugates) but due to different topology heparin seems to masks the RBD domain (yellow circle) and probably this is the reason for heparin (Unbound heparan sulfate) blocking spike-Ace2 interactions as also reported earlier [13]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
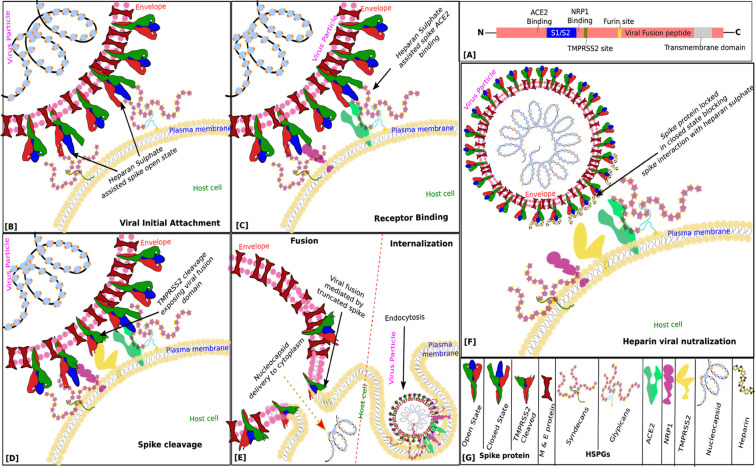
Fig. 5.
Cartoon representation of the proposed mechanism of heparan sulfate and heparin neutralization of SARS-CoV-2 virion. (A) This is a topological map of the domain architecture of spike protein showing different functional regions. (B) Based on the essential role of Heparan sulfate [17] and the docking results suggest an open state of spike protein upon interacting with HSPGs. (C) Interactions of open state spike protein with ACE2 and/or NRP1 receptors. (D) Cleavage of a spike by TMPRSS2 exposing viral fusion domains. (E) Fusion: Spike protein-mediated fusion of viral envelope with host plasma membrane resulting in delivery of nucleocapsid to host cell cytoplasm, Internalization: Formation of endosomes like inclusions mediated by Spike-Heparan sulfate-ACE2/NRP1 interactions leading to nucleocapsid delivery to host cytoplasm through further processing of spike by Furin and/or Cathepsin L and Viral envelope and endosomal membrane fusion similar to the plasma membrane.
The mutation mapping with respect to the heparin-binding site revealed a high conservancy of the site as all the mutations were lying away from the predicted interacting site (Fig. 6). These findings highlight the importance of heparan sulfate interactions in SARS-CoV-2 viral invasions as well as conserved inhibitory/neutralization potential of the free heparin sulfate against evolved strains with mutant forms of spike protein trimer.
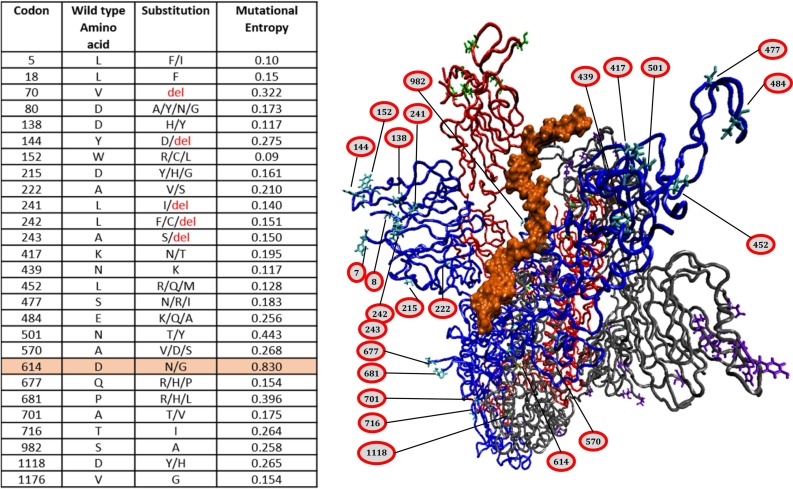
Fig. 6.
Relative mutation site analysis with respect to heparin binding to the closed state. Inset table lists the mutations reported as of February 2021 (GISAID-hcov-19 with entropy greater than 0.1). The three peptides constituting the spike trimer are colored red, blue and gray. Heparin mostly interacts with blue peptide in this pose, therefore, all the mutant positions are labelled on the same to avoid confusion. Interestingly most of the mutant locations are away from the heparin binding site which suggests important and highly conserved role of sulfated polysaccharides in the viral invasion as well as usability of heparin as a universal blocker of all the mutant spike variants. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Similar analysis with mutants revealed the binding and scores were comparable to the wild type we studied earlier (7CAI) (Fig. 7). We did find lower scores for Danish-Mink mutant predicted structures (−990). We cannot determine these differences are due to either mutations or the variability in the open state of the variants. There is a further need to evaluate the same through neutralization assays with the strains carrying spike polymorphisms.
Fig. 7.
Molecular docking analysis of heparin against spike protein (Closed state RBD) of SARS-CoV-2 envelope with different mutations. Position/location of the heparin molecule is pointed out with a circular purple ligand symbol. The trimeric protein complex had three mutant structures from PDB (7AD1, 7NDB, and 7KDL) and supercomputer predicted structures for mutants from UK, Brazil, South Africa, and Danish minks with differences in vaccine efficacies reported recently (Table 3). All the structure showed strong binding with closed state RBD. Lower docking score observed with Danish mink might not be necessarily be due to mutation but could be due to flexible state of the relatively large trimeric complex and also predicted structures might lack proper glycosylation's that seem to play an important role in spike stabilizations as seen in wild type structure 7CAI. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. In-vitro antiviral evaluation of heparin
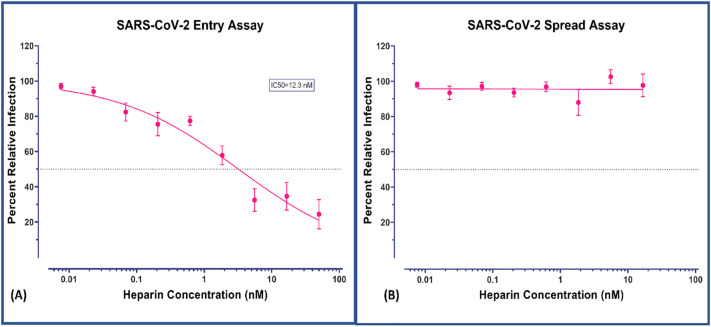
Following our virtual screening analysis of heparin, we performed in-vitro validation studies to confirm our computational results. The compound was tested for both entries of the virus to the host cells as well as spread to new cells upon infection. The results of the entry assay strongly support the specificity of the heparin against spike protein at nanomolar efficacy (IC50 = 12.3 nM/12300 pM, Fig. 2A). These results reiterate the potential neutralizing property of heparin if used as an entry blocker. Our study further confirms the current breadth of knowledge that suggests that heparin may have antiviral properties. We suspect that due to variations in testing procedures, previous studies have underreported the antiviral efficacy of heparin (Table 1 ). The reported efficacies of heparin/analogs against other viruses are included in Table 2 . In terms of the viral spread assay, the negative result in nanomolar range is not unexpected. This is because this assay detects viral progression primarily with major intracellular loads and therefore needs higher concentration (Table 1). As the agent is tested with already infected (seeded) cultures the MOI increases exponentially even before the next invasion cycle. Such numbers are not controlled by an invasion inhibitor at the tested concentration range. This also confirms the usefulness of specific entry assays for complete viruses to determine neutralization efficacy. Any therapeutic agents that interfere with the specific entry mechanism of SARS-CoV-2 will have contrasting results between these two assays. Our results demonstrate that dose-dependent and highly efficacious blocking of virus loads through the entry assay makes heparin an excellent candidate for transmission-blocking of SARS-CoV-2. (See Fig. 3.)
Fig. 2.
Evaluation of antiviral activities of heparin. (A) against virus attachment and entry/fusion. The experimental procedure, virus concentration (PFU/well or MOI), and the time of addition and treatment with the test compounds are presented in the method sections. (B) Nonobservance of inactivation of viral infections by the test compounds heparin. A virus surface spike protein mediates SARS-CoV-2 entry into cells. Our results are in agreement with previous reports and show the contrast in antiviral activity (Ic50 = 12.3 nM in entry assay vs. nil activity in spread assay in given concentration range) in different formats confirming the mechanism of action of heparin.
Table 1.
The reported efficacy of heparin against SARS-CoV-2. Note: N/A (not applicable).
| Type of heparin | Assay format | Efficacy | Proposed usage | Follow up trial (reference) | References | Comments |
|---|---|---|---|---|---|---|
| UFH | SARS-CoV-2 viral plaque forming assay | 70% inhibitio;100 μg/mL | Therapeutic | N/A | [16] | Recommendation as first-line use |
| “Pixatimod (PG545)” | Plaque assay, percent inhibition from the cytopathic effect | 32–51% inhibition; 100 μg/mL | Prophylactic/therapeutic | N/A | [22] | Emphasizes multi-modality if heparin; possibly as prophylactic and therapeutic |
| UFH | Pseudotyped Lentivirus entry inhibition | 5.99 μg/L | N/A | N/A | [32] | |
| UFH (formulation) | plaque inhibition assay with Vero E6 cells | 25–41 μg ml | Treatment | N/A | [33] | |
| Hexa- and octasaccharides | Surface plasma resonance (SPR) spike-heparin interaction inhibition | 38 nM | Therapeutic | [34] | [35] | Oligosaccharide based inhibitors |
| UFH (High mol weight fractions) | Virus binding assay to Calu3, human lung cancer cell line | <0.05 U/ml | Therapeutic | Nebulized heparin [36] | [37] | Long-chain heparin molecules are more effective than small ones` |
Table 2.
The reported efficacy of heparin against major non-SARS-CoV-2 viral pathogens. Note:-N/A (not applicable). Many other viruses are sensitive to heparin & sulfated polysaccharides [38].
| Type of heparin | Assay format | Efficacy | Proposed usage | Pathogen | References | Comments |
|---|---|---|---|---|---|---|
| UFH | Cellular cytopathogenicity | 50% cytopathogenic reduction at 7.5 μg/ml | Therapeutic | HIV | [39,40] | No toxicity at a concentration up to 400 μg/ml |
| Various forms of heparin | plaque inhibition assay and viral reduction assay with Vero cells | NA | NA | ZIKV | [41] | Promoted ZIKV replication. The longer the heparin chain, the stronger the replication promotion |
| Various forms of heparin | plaque inhibition assay and viral reduction assay with Vero cells | Up to 93.5% inhibition at 200 μg/ml | Therapeutic | DENV | [42,43] | All formulations of heparin used in the study inhibited DENV replication |
| Biomimetic heparins | Plaque inhibition assay with vero and 293TT cells | IC50 varied from 0.50–2.88 μg/ml | Therapeutic | HSV; HPV; RSV; ROTV | [44] | All of the studied viruses are heparan sulfate dependent |
3.3. Proposed heparin formulations for topical applications
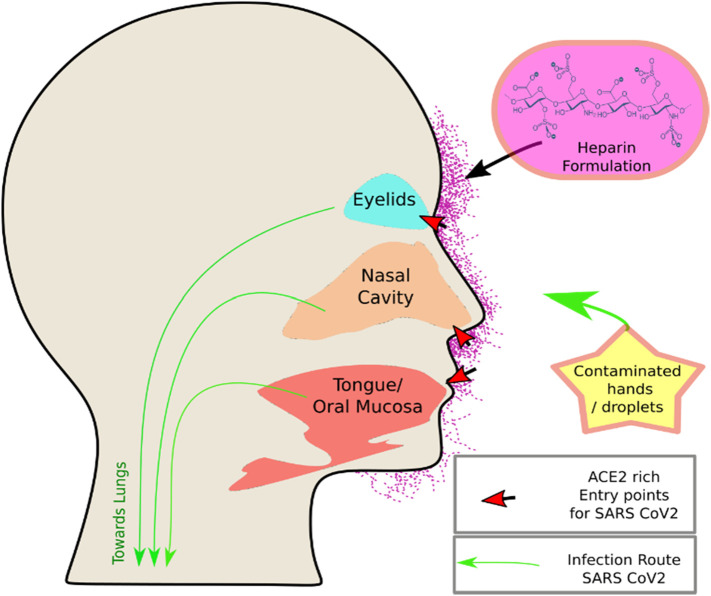
Although the anticoagulant activity of heparin has been extensively studied, studies show that UFH exhibits a varying level of biological activities due to its differential interaction characteristics with various proteins including antiviral, anti-inflammatory, and cytoprotective effects [[61], [62], [63]]. Sulfated polysaccharides in general have been shown to be inhibiting a number of viruses [38,64]. For example, various studies show that UFH has antiviral activity against many viruses causing major diseases which have cellular entry dependency on Heparan sulfate (Table 2). Spike protein of SARS-CoV-2 in particular has been reported to be highly susceptible to various other sulfated polysaccharides [18,65]. UFH is composed of a mixture of different heparan sulfate polysaccharides with a variety of target proteins in the host including growth factors. The topical use of heparin is a safer option compared to the systemic delivery [66], however, the transdermal penetration of heparin is demonstrated to be very low due to its large size (3–30 kDa), hydrophilicity, and negative charge. The local use of heparin on the skin produces some desired effects, including its anticoagulating and microcirculatory-modulatory actions as well as its assistance in facilitating the diffusion of other useful drugs through skin barriers [67,68]. However, a heavy dose of heparin does not necessarily improve penetration through the intact skin but on the contrary, it is on the verge of becoming a concern due to pertinent adverse effects [31]. Therefore, heparin, if used in the form of an effective formulation, could be a better choice due to the rational minimization of any possible side effects, competent skin penetration, and the consequent lowering of effective dose. In this context, liposomes, oil-in-water emulsions, microemulsions, viscous gels, pastes and creams, and sprayable formulations are usually developed to address these issues [[69], [70], [71]]. However, the most desirable feature of the formulation is the absorption of heparin in the stratum corneum barrier and consequent therapeutic effects. Heparin formulation that meets these requirements will have far greater chances of success specifically, for COVID-19 prevention. We are currently pursuing the lipid-based formulations as they are much more promising due to their indispensable properties, including the biocompatibility with skin lipids, its GRAS grade characters, amphiphilic nature enabling the efficacious loading, penetration, and delivery of heparin across the skin barriers attaining optimum therapeutic effect with minimal side effects [70,71]. Skin softening, operative moisturization, and impermeable film formation preventing an entry of undesired environmental and biochemical particles are other advantages provided by potential lipid-based nano-formulations. As such, our immediate focus is to develop a range of formulations and evaluate their applicability as translational blockers for COVID-19. These formulations will provide an ultimate platform in developing treatments against COVID-19, which is a necessity of the current unprecedented times. These formulations can be applied at the proven entry sites on the human face, namely the area near the eyes, nose, ears, and mouth as shown in Fig. 1 . Such a well-absorbed formulation can also be exploited for the control of other diseases, for example, HIV transmission-blocking as well as conventional usage of heparin for varicose vein/hemorrhoids venous thrombosis.
Fig. 1.
ACE2 rich Mucocutaneous tissue and upper respiratory mucosa are the entry points for the SARS-CoV-2 virus. The proposed intervention with heparin formulation should be the key to block transmission.
3.3.1. The safety profile of heparin
A strong safety profile and critical therapeutic applications have made heparin one of the most widely used clinical compounds in the world [2]. For most cases, the use of heparin at the recommended doses does not threaten the livelihood of the patient and several clinical studies have confirmed its benefit over minimal side effects [31]. Though, it must be understood that in special populations, especially those that have a high risk of bleeding, the benefit of using heparin must be critically evaluated before considering administration.
4. Conclusion and perspective
Our antiviral data strongly shows UFH can block viral entry at sub-nanomolar concentrations. Additionally, in-silico analysis suggesting free heparin interferes with cell surface heparan sulfate-Spike protein interaction, a critical part required for binding with ACE2 receptor (Fig. 4, Fig. 5). With the unabated spread of SARS-CoV-2, containment is the best strategy to mitigate this pandemic, although various lockdowns and social distancing practices are based on the same strategy. Additional measures should include lowering infection loads to exposure levels to aid in building herd immunity, such as wearing face masks and eye shields. To supplement these mitigation approaches, specifically with frontline workers, close contacts and highly vulnerable populations, we propose the potential application of heparin-based nasal sprays and mucocutaneous tropical formulations. Nebulized UFH has been proposed as a therapeutic agent against COVID-19 [36]. Preliminary reports show limited success with time reduction (duration of hospitalization) in extubating in COVID-19 patients [72]. Topical or nasal formulated applications with enhanced absorption may be a key to protect from highly contagious SARS-CoV-2 infection as the viral load stays minimal during the transmission stage of the infection cycle. The current topical applications are marred by only 4.2% bioavailability for subdermal thrombosis [73] and the dermal accumulation data is still unavailable. Therefore, there is ample scope for improving its efficacy through more effective formulations.
CRediT authorship contribution statement
YG and PK conceived and designed the HTVS study and performed in-silico analysis. YG and DM created the cartoons depicting the role of spike protein and heparin during viral entry. SEZ, ASH, and JMD performed in-vitro testing of top hits against the SARS-CoV-2 virus and cytotoxicity. CVK contributed the sections on heparin and its formulations. DM, RD, and JF contributed to the clinical aspect of COVID-19 and therapeutic applications of heparin. All authors contributed to writing and reviewing the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
The authors sincerely thank the Department of Medicine, Loyola University Medical Center, and Stritch School of Medicine for supporting the Drug Discovery Program and the High Computing Platform. This project was also supported by the Research Funding Committee (RFC), Loyola University Chicago (LU#213627). Funding for USAMRIID was provided through the CARES Act with programmatic oversight from the Military Infectious Diseases Research Program–project 14066041. Opinions, discussions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army. The mention of trade names or commercial products does not constitute endorsement or recommendation for use by the Department of the Army or the Department of Defense.
References
- 1.Hill A.M., Barber M.J., Gotham D. Estimated costs of production and potential prices for the WHO Essential Medicines List. BMJ Glob. Health. 2018;3 doi: 10.1136/bmjgh-2017-000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization WH, et al. 2019. The Selection and Use of Essential Medicines: Report of the WHO Expert Committee on Selection and Use of Essential Medicines, 2019 (Including the 21st WHO Model List of Essential Medicines and the 7th WHO Model List of Essential Medicines for Children) [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arachchillage D.R., Laffan M. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18:1233–1234. doi: 10.1111/jth.14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020;18(5):1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lillicrap D. Disseminated intravascular coagulation in patients with 2019-nCoV pneumonia. J. Thromb. Haemost. 2020;18:786. doi: 10.1111/jth.14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood J. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong M., Liang X., Wei Y.-D. Changes in blood coagulation in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Br. J. Haematol. 2020;189(6):1050–1052. doi: 10.1111/bjh.16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Li Y., Yang B., Wang H., Li L. Low-molecular-weight heparin treatment for acute lung injury/acute respiratory distress syndrome: a meta-analysis of randomized controlled trials. Int. J. Clin. Exp. Med. 2018;11:414–422. [Google Scholar]
- 12.Shi C., Wang C., Wang H., Yang C., Cai F., Zeng F., et al. The potential of low molecular weight heparin to mitigate cytokine storm in severe COVID-19 patients: a retrospective cohort study. Clin. Transl. Sci. 2020;13(6):1087–1095. doi: 10.1111/cts.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thachil J. The versatile heparin in COVID-19. J. Thromb. Haemost. 2020;18:1020–1022. doi: 10.1111/jth.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Testa S., Paoletti O., Giorgi-Pierfranceschi M., Pan A. Switch from oral anticoagulants to parenteral heparin in SARS-CoV-2 hospitalized patients. Intern. Emerg. Med. 2020:1–3. doi: 10.1007/s11739-020-02331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajra A., Mathai S.V., Ball S., Bandyopadhyay D., Veyseh M., Chakraborty S., et al. Management of thrombotic complications in COVID-19: an update. Drugs. 2020:1–10. doi: 10.1007/s40265-020-01377-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mycroft-West Courtney J., Su Dunhao, Pagani Isabel, Rudd Timothy R., Elli Stefano, Gandhi Neha S., Guimond Scott E., et al. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin. Throm. Haemos. 2020;120(12):1700–1715. doi: 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge L.J., Urwin L., Nicklin M.J., James D.C., Green L.R., Monk P.N. ACE2-independent interaction of SARS-CoV-2 spike protein to human epithelial cells can be inhibited by unfractionated heparin. BioRxiv. 2020 doi: 10.1101/2020.05.21.107870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon P.S., Oh H., Kwon S.-J., Jin W., Zhang F., Fraser K., et al. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AB F.B., Sarialioglu F. The old but new: can unfractioned heparin and low molecular weight heparins inhibit proteolytic activation and cellular internalization of SARS-CoV2 by inhibition of host cell proteases? Med. Hypotheses. 2020:109743. doi: 10.1016/j.mehy.2020.109743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Miller S., Patek M., Moutal A., Cabel C.R., Thorne C.A., Campos S.K., et al. In silico identification and validation of inhibitors of the interaction between neuropilin receptor 1 and SARS-CoV-2 spike protein. Biorxiv. 2020 doi: 10.1101/2020.09.22.308783. [DOI] [Google Scholar]
- 21.Bermejo-Jambrina M., Eder J., Kaptein T.M., Helgers L.C., Brouwer P.J., van Hamme J.L., et al. SARS-CoV-2 infection and transmission depends on heparan sulfates and is blocked by low molecular weight heparins. BioRxiv. 2020 doi: 10.1101/2020.08.18.255810. [DOI] [Google Scholar]
- 22.Guimond S.E., Mycroft-West C.J., Gandhi N.S., Tree J.A., Buttigieg K.R., Coombes N., et al. Synthetic heparan sulfate mimetic pixatimod (PG545) potently inhibits SARS-CoV-2 by disrupting the spike-ACE2 interaction. BioRxiv. 2020 doi: 10.1101/2020.06.24.169334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta Y., Maciorowski D., Zak S.E., Jones K.A., Kathayat R.S., Azizi S.-A., et al. Bisindolylmaleimide IX: a novel anti-SARS-CoV2 agent targeting viral main protease 3CLpro demonstrated by virtual screening pipeline and in-vitro validation assays. Methods. 2021 doi: 10.1016/j.ymeth.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta Y., Maciorowski D., Jones K., Kathayat R., Azizi S., Catherine P., et al. 2020. Bisindolylmaleimide IX: A Novel Anti-SARS-CoV2 Agent Targeting Viral Main Protease 3CLpro Demonstrated by Virtual Screening and In Vitro Assays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan S., Gor J., Mulloy B., Perkins S.J. Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin–protein complexes. J. Mol. Biol. 2010;395:504–521. doi: 10.1016/j.jmb.2009.10.064. [DOI] [PubMed] [Google Scholar]
- 27.Schrodinger L. Vol. 2020. 2020. Small-Molecule Drug Discovery Suite 2020–1. [Google Scholar]
- 28.Schrodinger P. Schrodinger LLC; N Y NY: 2020. Release 2020–2: Maestro. [Google Scholar]
- 29.Release S . Schrödinger LLC; N Y: 2020. 2: Schrödinger Suite 2020–2 Glide. [Google Scholar]
- 30.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alban S. Heparin- Century Prog. Springer; 2012. Adverse effects of heparin; pp. 211–263. [Google Scholar]
- 32.Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., et al. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J. Virol. 2021;95(3) doi: 10.1128/JVI.01987-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tree J.A., Turnbull J.E., Buttigieg K.R., Elmore M.J., Coombes N., Hogwood J., et al. Unfractionated heparin inhibits live wild type SARS-CoV-2 cell infectivity at therapeutically relevant concentrations. Br. J. Pharmacol. 2021;178(3):626–635. doi: 10.1111/bph.15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conzelmann C., Müller J.A., Perkhofer L., Sparrer K.M., Zelikin A.N., Münch J., et al. Inhaled and systemic heparin as a repurposed direct antiviral drug for prevention and treatment of COVID-19. Clin. Med. 2020;20:e218. doi: 10.7861/clinmed.2020-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L., Chopra P., Li X., Wolfert M.A., Tompkins S.M., Boons G.-J. SARS-CoV-2 spike protein binds heparan sulfate in a length-and sequence-dependent manner. BioRxiv. 2020 doi: 10.1101/2020.05.10.087288. [DOI] [Google Scholar]
- 36.Van Haren F.M., Page C., Laffey J.G., Artigas A., Camprubi-Rimblas M., Nunes Q., et al. Nebulised heparin as a treatment for COVID-19: scientific rationale and a call for randomised evidence. Crit. Care. 2020;24:1–11. doi: 10.1186/s13054-020-03148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Partridge L.J., Green L.R., Monk P.N. Unfractionated heparin potently inhibits the binding of SARS-CoV-2 spike protein to a human cell line. BioRxiv. 2020 doi: 10.1101/2020.05.21.107870. [DOI] [Google Scholar]
- 38.Kamhi E., Joo E.J., Dordick J.S., Linhardt R.J. Glycosaminoglycans in infectious disease. Biol. Rev. 2013;88:928–943. doi: 10.1111/brv.12034. [DOI] [PubMed] [Google Scholar]
- 39.Ito M., Baba M., Sato A., Pauwels R., De Clercq E., Shigeta S. Inhibitory effect of dextran sulfate and heparin on the replication of human immunodeficiency virus (HIV) in vitro. Antivir. Res. 1987;7:361–367. doi: 10.1016/0166-3542(87)90018-0. [DOI] [PubMed] [Google Scholar]
- 40.Harrop H.A., Coombe D.R., Rider C.C. Heparin specifically inhibits binding of V3 loop antibodies to HIV-1 gp120, an effect potentiated by CD4 binding. AIDS (London, England) 1994;8:183–192. doi: 10.1097/00002030-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 41.Kim S.Y., Koetzner C.A., Payne A.F., Nierode G.J., Yu Y., Wang R., et al. Glycosaminoglycan compositional analysis of relevant tissues in Zika virus pathogenesis and in vitro evaluation of heparin as an antiviral against Zika virus infection. Biochemistry. 2019;58:1155–1166. doi: 10.1021/acs.biochem.8b01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung S.-L., Lee P.-L., Chen H.-W., Chen L.-K., Kao C.-L., King C.-C. Analysis of the steps involved in dengue virus entry into host cells. Virology. 1999;257:156–167. doi: 10.1006/viro.1999.9633. [DOI] [PubMed] [Google Scholar]
- 43.Lin Y.-L., Lei H.-Y., Lin Y.-S., Yeh T.-M., Chen S.-H., Liu H.-S. Heparin inhibits dengue-2 virus infection of five human liver cell lines. Antivir. Res. 2002;56:93–96. doi: 10.1016/s0166-3542(02)00095-5. [DOI] [PubMed] [Google Scholar]
- 44.Lembo D., Donalisio M., Laine C., Cagno V., Civra A., Bianchini E.P., et al. Auto-associative heparin nanoassemblies: a biomimetic platform against the heparan sulfate-dependent viruses HSV-1, HSV-2, HPV-16 and RSV. Eur. J. Pharm. Biopharm. 2014;88:275–282. doi: 10.1016/j.ejpb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Sberna G., Amendola A., Valli M.B., Carletti F., Capobianchi M.R., Bordi L., et al. Trend of respiratory pathogens during the COVID-19 epidemic. J. Clin. Virol. 2020;104470:129. doi: 10.1016/j.jcv.2020.104470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zucman N., Uhel F., Descamps D., Roux D., Ricard J.D. Severe reinfection with South African SARS-CoV-2 variant 501Y.V2: a case report. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.2021. Risk Assessment: Risk Related to the Spread of New SARS-CoV-2 Variants of Concern in the EU/EEA—First Update.https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update Available online: [Google Scholar]
- 48.WHO WHO SARS-CoV-2 variant—United Kingdom of Great Britain and Northern Ireland. https://www.who.int/csr/don/21-december-2020-sars-cov2-variant-united-kingdom/en.
- 49.Fiorentini S., Messali S., Zani A., Caccuri F., Giovanetti M., Ciccozzi M., et al. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang J.W., Toovey O.T., Harvey K.N., Hui D.D. Introduction of the South African SARS-CoV-2 variant 501Y. V2 into the UK. J. Inf. Secur. 2021 doi: 10.1016/j.jinf.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., et al. SARS-CoV-2 501Y. V2 escapes neutralization by south African COVID-19 donor plasma. Nat. Med. 2021:1–4. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 52.2020. European Centre for Disease Prevention and Control, Rapid Risk Assessment: Detection of new SARS-CoV-2 variants related to mink, ECDC, Stockholm.www.ecdc.europa.eu/en/publications-data/detection-new-sars-cov-2-variants-mink [Google Scholar]
- 53.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 54.Voloch C.M., da Silva Francisco R., de Almeida L.G., Cardoso C.C., Brustolini O.J., Gerber A.L., et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95(10):e00119–e00121. doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faria N.R., Claro I.M., Candido D., Moyses Franco L., Andrade P.S., Coletti T.M., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2021 https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 [Google Scholar]
- 56.Resende P.C., Bezerra J.F., de Vasconcelos R.H.T., Arantes I., Appolinario L., Mendonça A.C., et al. Vol. 10. January 2021. Spike E484K Mutation in the First SARS-CoV-2 Reinfection Case Confirmed in Brazil, 2020; p. 2021. [Google Scholar]
- 57.New Variant Strain of SARS-CoV-2 Identified in Travelers from Brazil. National Institute of Infectious Diseases; JAPAN: 2021. [Google Scholar]
- 58.Juraszek J., Rutten L., Blokland S., Bouchier P., Voorzaat R., Ritschel T., et al. Stabilizing the closed SARS-CoV-2 spike trimer. Nat. Commun. 2021;12:1–8. doi: 10.1038/s41467-020-20321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gobeil S., Janowska K., McDowell S., Mansouri K., Parks R., Manne K., et al. D614G mutation alters SARS-CoV-2 spike conformational dynamics and protease cleavage susceptibility at the S1/S2 junction. BioRxiv. 2021;34(2):108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M., Supasa P., Case J.B., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young E. The anti-inflammatory effects of heparin and related compounds. Thromb. Res. 2008;122:743–752. doi: 10.1016/j.thromres.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 62.Li J.-P., Vlodavsky I. Heparin, heparan sulfate and heparanase in inflammatory reactions. Thromb. Haemost. 2009;102:823–828. doi: 10.1160/TH09-02-0091. [DOI] [PubMed] [Google Scholar]
- 63.Shukla D., Spear P.G., et al. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S.Y., Jin W., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir. Res. 2020;104873:181. doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin W., Zhang W., Mitra D., McCandless M.G., Sharma P., Tandon R., et al. The structure-activity relationship of the interactions of SARS-CoV-2 spike glycoproteins with glucuronomannan and sulfated galactofucan from Saccharina japonica. Int. J. Biol. Macromol. 2020;163:1649–1658. doi: 10.1016/j.ijbiomac.2020.09.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pandya J.M, Gupta S., Chouhan A., et al. Evaluation of Safety and Efficacy of Quick Penetrating Heparin Solution (1000 IU/ml) in Prevention of Intravenous Cannula Related Thrombophlebitis: A Prospective, Randomized, Comparative, Parallel Group Clinical Study. Indian J Anesth Analg. 2019;6(6 Part -II):2129–2132. [Google Scholar]
- 67.Bonina F., Montenegro L. Effects of some non-toxic penetration enhancers on in vitro heparin skin permeation from gel vehicles. Int. J. Pharm. 1994;111:191–196. [Google Scholar]
- 68.Cesarone M., Belcaro G., Errichi S., Cornelli U., Pellegrini L., Ruffini I., et al. Topical heparin: new observations. Angiology. 2007;58:16S–20S. doi: 10.1177/0003319706297740. [DOI] [PubMed] [Google Scholar]
- 69.Belcaro G., Cesarone M., Dugall M., Feragalli B., Ippolito E., Corsi M., et al. Topical formulation of heparin is effective in reducing the symptoms of superficial venous thrombosis: a monocenter, observer-blind, placebo-controlled randomized study. Panminerva Med. 2011;53:3–11. [PubMed] [Google Scholar]
- 70.Kulkarni C.V. Lipid self-assemblies and nanostructured emulsions for cosmetic formulations. Cosmetics. 2016;3:37. [Google Scholar]
- 71.Kulkarni C.V., Vishwapathi V.K., Quarshie A., Moinuddin Z., Page J., Kendrekar P., et al. Self-assembled lipid cubic phase and cubosomes for the delivery of aspirin as a model drug. Langmuir. 2017;33:9907–9915. doi: 10.1021/acs.langmuir.7b02486. [DOI] [PubMed] [Google Scholar]
- 72.Dixon B., Smith R., Artigas A., Laffey J., McNicholas B., Schmidt E., et al. Can nebulised heparin reduce time to extubation in SARS CoV 2 the CHARTER study protocol. MedRxiv. 2020 doi: 10.1101/2020.04.28.20082552. [DOI] [Google Scholar]
- 73.Vecchio C., Frisinghelli A. Topically applied heparins for the treatment of vascular disorders. Clin. Drug Investig. 2008;28:603–614. doi: 10.2165/00044011-200828100-00001. [DOI] [PubMed] [Google Scholar]
- 74.Weisshoff H., Krylova O., Nikolenko H., Düngen H.-D., Dallmann A., Becker S., et al. Aptamer BC 007-efficient binder of spreading-crucial SARS-CoV-2 proteins. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05421. [DOI] [PMC free article] [PubMed] [Google Scholar]