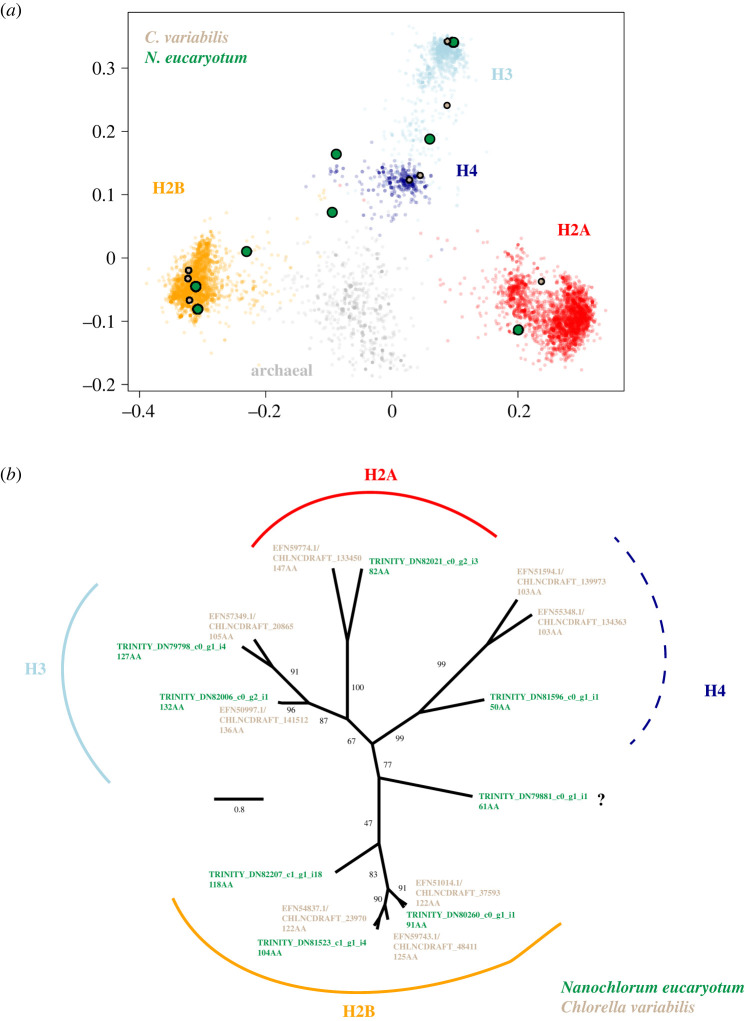
Figure 1.
Nanochlorum eucaryotum (SAG 55.87) candidate histones in phylogenetic context. (a) Similarity of eukaryotic and archaeal histones based on classic multidimensional scaling, which renders distance relationships (derived from the alignment) in two dimensions. Histones annotated as H2A, H2B, H3, H4 or of archaeal origin are represented by distinct colours and fall into reasonably distinct clusters. Histones from C. variabilis and histone candidates from N. eucaryotum (SAG 55.87) associate with each of the eukaryotic clusters, suggesting that orthologues for all four core histones are present in these species. The alignment underlying this analysis is provided in Stevens et al. [22]. (b) Maximum-likelihood phylogenetic tree of C. variabilis and N. eucaryotum (SAG 55.87) histones. The length of the relevant polypeptides is provided in amino acids (AA) for each sequence. Nanochlorum eucaryotum (SAG 55.87) candidate histones are represented by IDs from the de novo transcript assembly (see electronic supplementary material, table S1). A single reconstructed peptide whose affiliation to either H4 or H2B is uncertain is marked with a question mark.