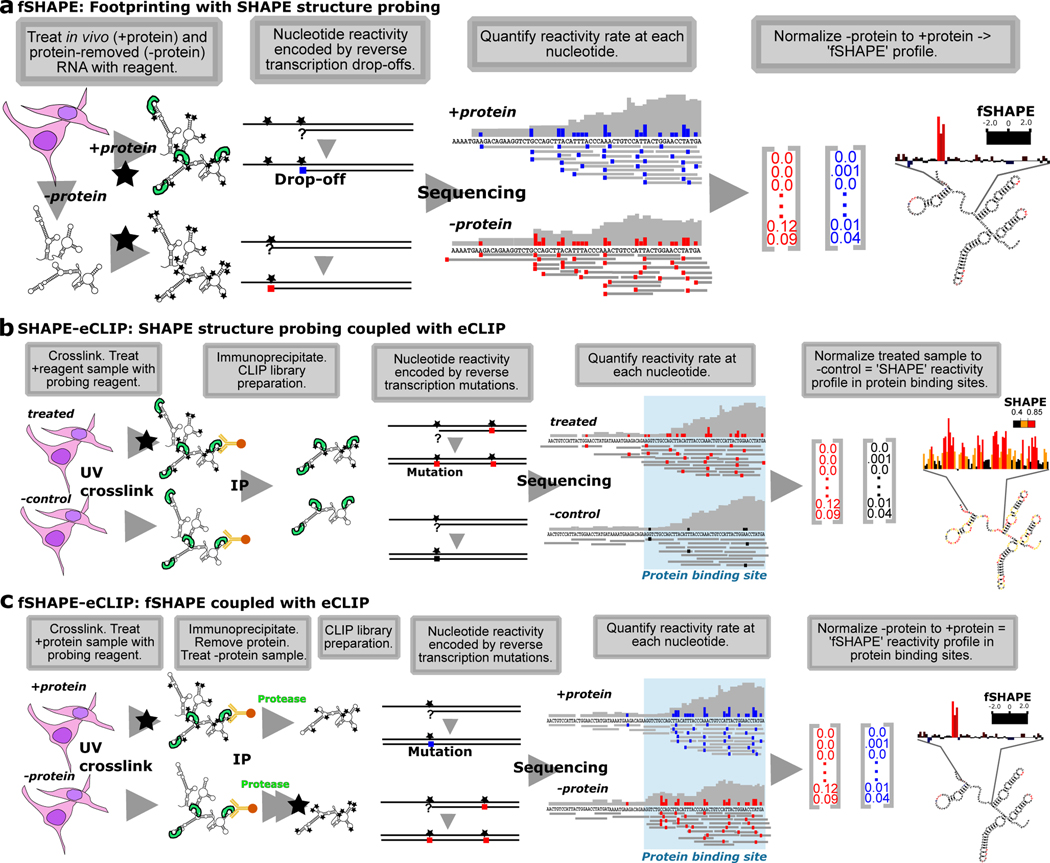
Figure 1.
SHAPE-based technologies for footprinting and structure probing of RNA-protein interfaces. (a) fSHAPE requires two RNA samples processed in parallel: a “+protein” samples in which cellular RNA is treated with the probing reagent (black star) and a “-protein” sample in which RNA is extracted from cells, stripped of protein, and treated with the probing reagent. Nucleotides that react with the reagent form adducts that result in drop-off events (colored square) during reverse transcription, such that the frequency of drop-off events at a given nucleotide is proportional to its reactivity rate with the probing reagent. “+protein” drop-off frequencies are subtracted from “-protein” drop-off frequencies and normalized to obtain an fSHAPE reactivity value at each nucleotide describing its degree of increased reactivity with the reagent in the absence of protein, akin to footprinting. (b) SHAPE-CLIP probes secondary structure in transcripts selected by CLIP. Cell samples are either treated with a structure probing reagent (black star) or an untreated negative control sample. Samples are UV crosslinked and extracted protein-bound transcripts are immunoprecipitated (IP) with an antibody to the desired protein. Nucleotides that react with the reagent form adducts that result in mutations (colored square) during a modified reverse transcription (Siegfried et al., 2014), such that the frequency of sequenced mutations at a given nucleotide is proportional to its reactivity rate with the probing reagent. “Treated” sample mutation rates are subtracted from “-control” mutation rates and normalized to obtain a SHAPE reactivity value at each nucleotide. Sequencing reads are also be used to determine protein binding sites (Van Nostrand et al., 2016). (c) fSHAPE-CLIP identifies nucleotides bound by protein in transcripts selected by CLIP. Cell samples are either initially treated with a structure probing reagent (“+protein”) or untreated (“-protein”). Samples are UV crosslinked and extracted protein-bound transcripts are immunoprecipitated (IP) with an antibody to the desired protein. RNA is protease-treated and refolded; the “-protein” samples is treated with the structure probing reagent. Nucleotides that react with the reagent form adducts that result in mutations (colored square) during a modified reverse transcription (Siegfried et al., 2014), such that the frequency of sequenced mutations at a given nucleotide is proportional to its reactivity rate with the probing reagent. “+protein” sample mutation rates are subtracted from “-protein” mutation rates and normalized to obtain an fSHAPE reactivity value at each nucleotide. Sequencing reads are also be used to determine protein binding sites (Van Nostrand et al., 2016). See also Table S1.