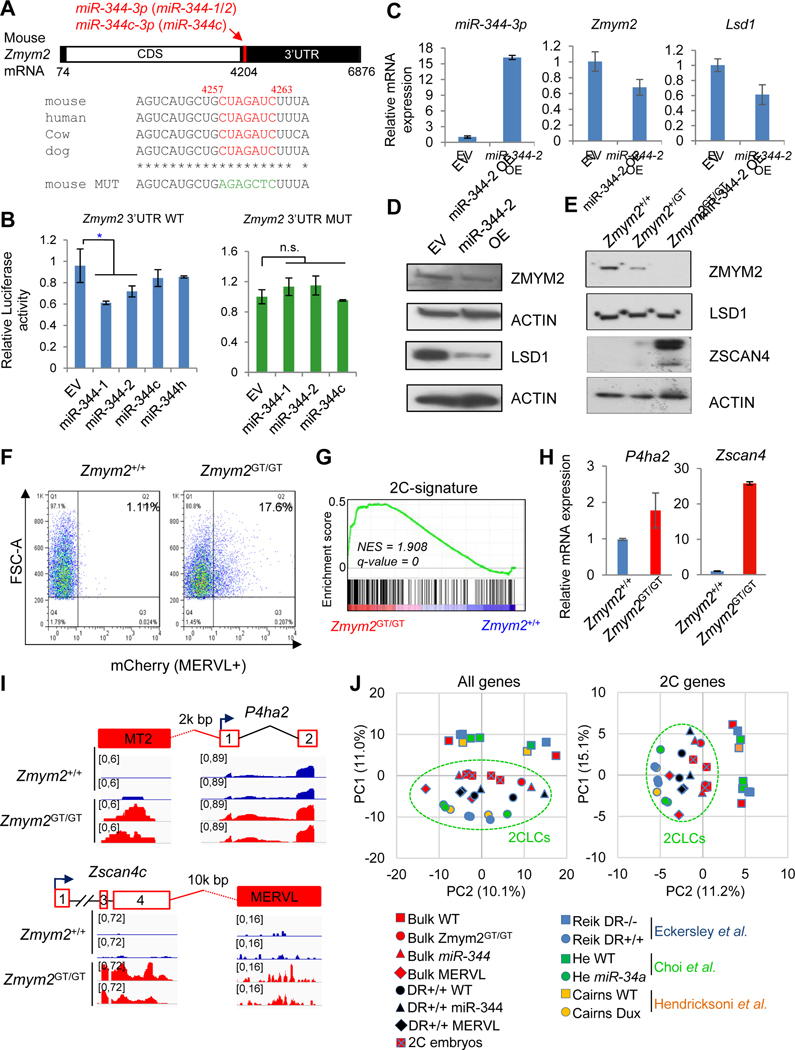
Figure 4. Zmym2 and Lsd1 are the targets of miR-344.
(A) Schematic layout of the Zmym2 mRNA with predicted miR-344 binding sites indicated. The evolutionarily conserved seed sequence and its mutated version are indicated.
(B) Luciferase reporter assay in ESCs co-transfected with reporter constructs containing the Zmym2 WT (left) or mutated (right) 3’UTRs and miR-344-1/2/c/h expression vectors or empty vector (EV). miR-344-h is a non-targeting control. Data are presented as average ± SD, P-value is from T-test, * P<0.05, n.s. non-significant.
(C) Relative expression levels of miR-344-3p, Zmym2, and Lsd1 upon overexpression (OE) of miR-344-2 or empty vector (EV). RT-qPCR data are presented as average ± SD.
(D) Protein expression of ZMYM2 and LSD1 in mESCs after overexpression (OE) of miR-344-2 or empty vector (EV). ACTIN is a loading control.
(E) Protein expression of ZMYM2, LSD1, and ZSCAN4 in Zmym2+/+, Zmym2+/GT, Zmym2GT/GT ESCs. ACTIN is a loading control.
(F) FACS profiles of 2C::tdTomato positive cells in Zmym2+/+ and Zmym2GT/GT ESCs.
(G) GSEA indicating that upregulated genes in Zmym2GT/GT ESCs were highly enriched in the 2-cell embryo gene set. Red, upregulated genes; blue, downregulated genes.
(H) RT-qPCR of 2C-specific P4ha2 and Zscan4c expression in Zmym2+/+ and Zmym2GT/GT ESCs. Data are presented as average ± SD.
(I) Examples of induced expression (shown as RNA-seq peaks) of MERVL-proximal P4ha2 and Zscan4c in Zmym2GT/GT compared with Zmym2+/+ cells.
(J) Principle-component analysis (PCA) of RNA-seq data from bulk or sorted DR+/+ population of miR-344-/MERVL-activated cells, Zmym2GT/GT ESCs, 2C embryos, and other published 2CLC cells. PCA was performed based on all genes (left) and only 2C-specific genes (Macfarlan et al., 2012) (right).