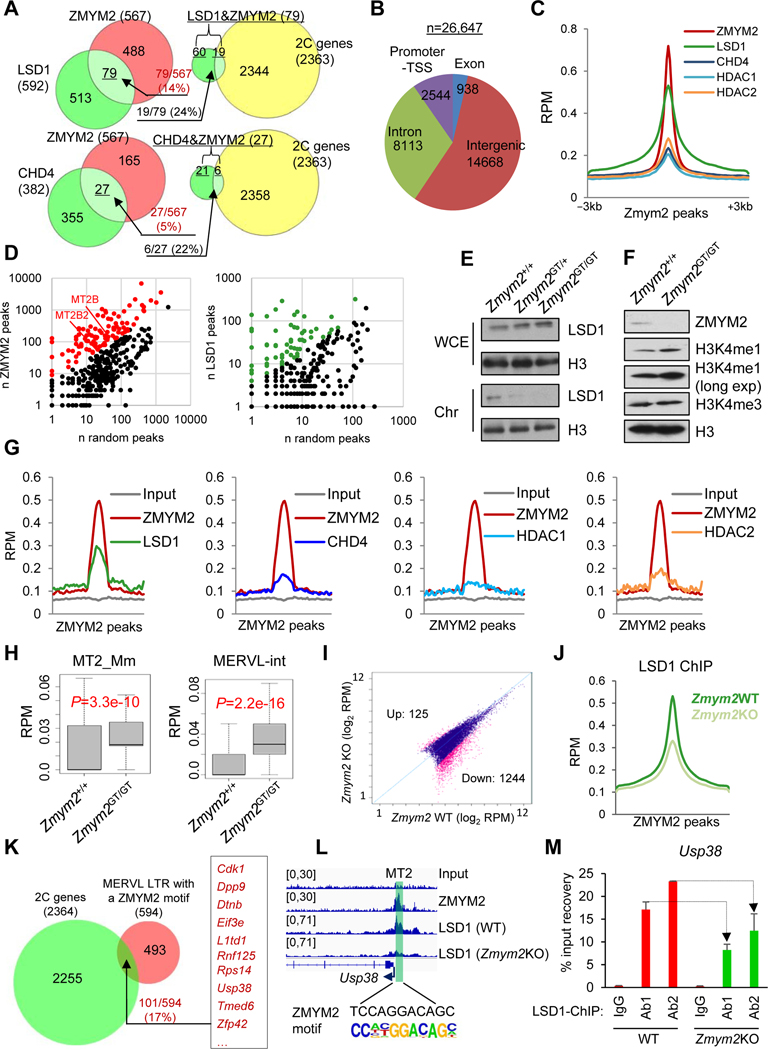
Figure 5. ZMYM2 physically associates and functionally interacts with the LSD1-NuRD corepressor complex in controlling MERVL expression.
(A) Overlap of the common genes repressed by ZMYM2/LSD1 (top) or ZMYM2/CHD4 (bottom), which are compared with the 2C-specific genes (Macfarlan et al., 2012).
(B) Distribution of ZMYM2 ChIP-seq peaks.
(C, G and J) Average ChIP-seq density (reads per million, RPM) of factors indicated around the ZMYM2 peak center (−3 kb to 3 kb) (C and J) or at ZMYM2 peaks overlapped with MERVL LTR (MT2) regions (G).
(D) Relative enrichment of MERVL elements (MT2B and MT2B2) in ZMYM2 (left) but not in LSD1 (right) peaks regions over a random control.
(E-F) Reduced chromatin occupancy of LSD1 (E) and increased H3K4me1 expression (F) upon Zmym2 depletion. H3 is a loading control. WCE, whole cell extracts; Chr, chromatin-bound fractions.
(H) Expression (reads per million, RPM) of the MERVL solo LTR (MT2_Mm, left) and internal region (MERVL-int, right) in WT and Zmym2GT/GT ESCs. P-value is from Mann-Whitney test.
(I) LSD1 ChIP-seq peak intensity in WT versus Zmym2KO ESCs. Significantly decreased and increased numbers of peaks upon loss of ZMYM2 are indicated with FDR < 0.01.
(K) Overlap of genes activated in 2C embryos (n=2363) and MERVL-proximal genes with a ZMYM2 motif at LTR promoter (n=594).
(L) ZMYM2 and LSD1 cobind at Usp38 promoter with an MT2 region.
(M) LSD1 ChIP-qPCR with 2 different antibodies, at promoters of Usp38 in WT and Zmym2KO ESCs. Data are presented as average ± SD.