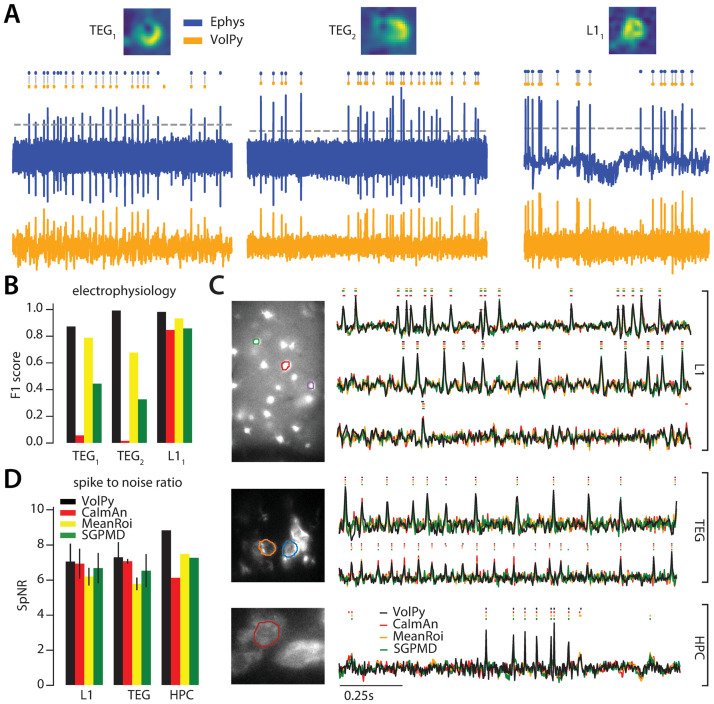
Fig 5. VolPy performance on real data.
(A) Evaluation of VolPy spike extraction performance against simultaneous electrophysiology. Three neurons, two from larval zebrafish TEG area (TEG1 and TEG2) and one from mouse L1 (L11), for which we had available both electrophysiology and imaging. Top. Spatial footprint extracted by VolPy. Middle. Ground truth spikes from electrophysiology (blue) and spikes extracted by VolPy (orange), gray vertical lines indicate matched spikes. Bottom. Electrophysiology (blue, top) and fluorescence signal denoised by VolPy (bottom, orange). (B) We compared the performance of VolPy, CaImAn, MeanROI, and SGPMD-NMF in retrieving spikes on the three neurons in (A). (C) Examples of trace extraction results for VolPy, CaImAn, MeanROI, and SGPMD-NMF. On the left mean image overlaid to example neurons (top L1, middle TEG, bottom HPC). On the right traces and inferred spikes for datasets L1 (top three traces), TEG (traces 4-5 from top) and HPC (bottom trace). (D) Spike to noise ratio (SpNR) for each considered algorithm and dataset type.