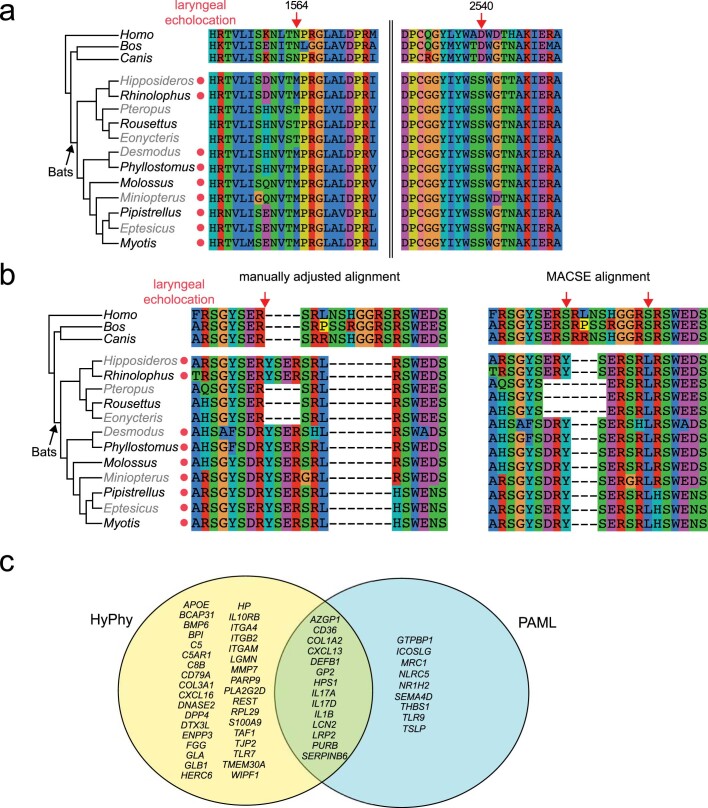
Extended Data Fig. 6. Screens for positive selection in genes in bats.
a, Sites under positive selection in bats in the LRP2 gene. Multiple sequence alignments of local regions surrounding two bat-specific mutations, which were found to be under positive selection (BEB > 0.95) using codeml (PAML). Site 1564 shows bat-specific changes at a conserved residue. The paraphyletic echolocating bats (indicated by a red dot) all share a methionine at this site, whereas pteropodid bats—which do not use laryngeal echolocation—have a threonine at this site. Site 2540 shows a bat-specific change, shared by all bats. The presence and patterns of mutations found in our six bats were confirmed in six previously published bat genomes, to increase taxonomic representation. Human (Homo), cow (Bos) and dog (Canis) are also shown. b, Echolocator-specific changes in the TJP2 gene. We initially identified positive selection in the bat ancestor in the hearing-related gene TJP2 (tight junction protein 2), which is expressed in cochlear hair cells and associated with hearing loss43. The right side shows the multiple sequence alignment produced by MACSE of local regions surrounding bat-specific mutations (red arrows), which were found to be under positive selection (BEB > 0.95) using codeml (PAML). The paraphyletic echolocating bats are indicated by a red dot. However, as shown on the left, manual inspection revealed a putative alignment ambiguity and manual adjustment produced an alignment with two bat-specific indels. This manually corrected alignment had a reduced significance for positive selection (aBSREL raw P = 0.009, not significant after multiple test correction considering 12,931 genes). The corrected alignment revealed a four-amino-acid microduplication found only in echolocating bats (n = 9) and not in pteropodid bats that lack laryngeal echolocation. This may be explained by incomplete lineage sorting or convergence. Insertions and deletions may also affect protein function, but are not considered by tests for positive selection; however, a phylogenetic interpretation of these events may uncover functional adaptations. c, Ageing and immune candidate genes showing evidence of significant positive selection using aBSREL (HyPhy, yellow) and codeml (PAML, blue). Genes identified by both methods are displayed at the intersection.