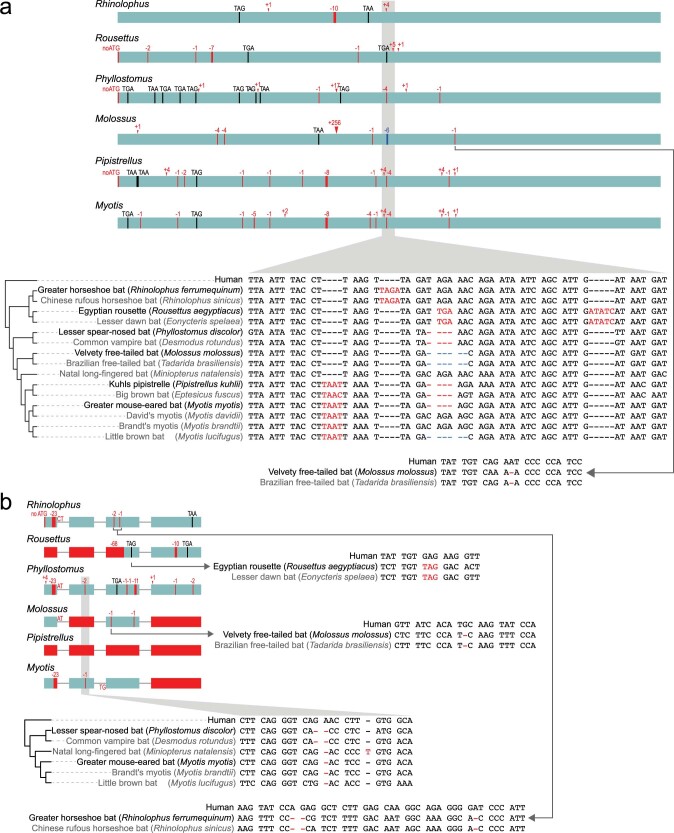
Extended Data Fig. 7. Inactivating mutations in LRRC70 and IL36G genes in bats.
a, b, LRRC70 (a) is expressed in a broad range of tissues and potentiates cellular responses to multiple cytokines54 and is well-conserved among Laurasiatheria. Importantly, LRRC70 strongly amplifies bacterial-lipopolysaccharide-mediated NF-κB activation54. Our finding of LRRC70 loss in bats makes this poorly characterized gene an interesting target for future mechanistic studies. IL36G (b) encodes a proinflammatory interleukin belonging to the interleukin-1 family. Increased expression of IL36G was detected in patients with psoriasis or inflammatory bowel disease, and IL36G is probably involved in the pathophysiology of these diseases by inducing the canonical NF-κB pathway and other proinflammatory cytokines55–57. Coding exons are represented as boxes (LRRC70 has only a single coding exon), superimposed with all detected inactivating mutations. Vertical red lines show frameshifting deletions; arrowheads indicate frameshifting insertions. Red boxes indicate complete or partial exon deletions. The size of deletions or insertions is given on top of the mutation. Premature stop codons are indicated by black vertical lines and the corresponding triplet. Mutated ATG start codons are indicated as ‘noATG’. Splice site mutations are shown by red letters at the end of an exon (donor mutation) or the beginning of an exon (acceptor mutation). One representative mutation for each bat is shown in detail in the alignment between human and bats (red font indicates the inactivating mutation). Genome assemblies produced in this study are in black; publicly available assemblies of sister species are in grey font. For both genes, the presence of the exact same mutation in independently sequenced and assembled genomes of sister species excludes the possibility that the representative mutations are erroneous. This analysis also reveals that both genes were in fact lost multiple times within Chiroptera, suggesting these genes came under relaxed selection in bats followed by with subsequent gene losses. In a, the position of the −4-bp frameshifting deletion in LRRC70 in Pipistrellus and Myotis is ambiguous and can be shifted by up to 3 bp to the right without affecting alignment identity.