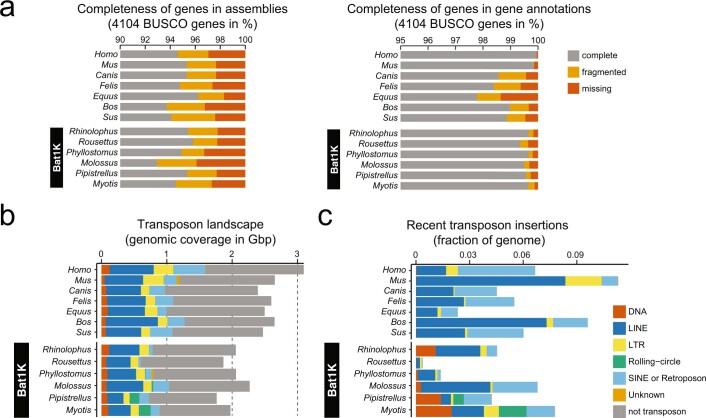
Extended Data Fig. 3. Comparison of assembly completeness in coding genomic regions and transposon content.
a, BUSCO applied to genomic sequences markedly underestimates gene completeness of assemblies. Bar charts show the percent of 4,104 highly conserved mammalian BUSCO genes that are completely present, fragmented or missing in the assembly. Left, applying BUSCO to genome assemblies. Right, applying BUSCO to the gene annotations (protein sequences of annotated genes; this panel is reproduced from Fig. 1d to enable a direct comparison). The direct comparison shows that BUSCO applied to the whole genome detects markedly fewer genes than BUSCO applied to the gene annotation. Because every annotated gene is by definition present in the assembly, this shows that BUSCO applied to the whole genome underestimates gene completeness—probably because it is substantially more difficult to detect complete genes in assemblies. b, Comparison of genomic transposon composition between six bats and other representative boreoeutherian mammals (Laurasiatheria + Euarchontoglires), selected for the highest genome contiguity. We used a previously described workflow and manual curation to annotate TEs82. Bar charts compare genome sizes and the proportion that consist of major transposon classes. TE content generally relates with genome size. Our assemblies also revealed noticeable genome size differences within bats, with assembly sizes ranging from 1.78 Gb for Pipistrellus to 2.32 Gb for Molossus. c, Fraction of the genome that consists of recent transposon insertions. We compared TE copies to their consensus sequence to obtain a relative age from each TE family. This revealed an extremely variable repertoire of TE families with evidence of recent accumulation (defined as TE insertions that diverged less than 6.6% from their consensus sequence). For example, while only about 0.38% of the 1.89-Gb Rousettus genome exhibits recent TE accumulations, about 4.2% of the similarly sized 1.78-Gb Pipistrellus genome is derived from recent TE insertions. The types of TE that underwent recent expansions also differ substantially in bats compared to other mammals, particularly with regards to the evidence of recent accumulation by rolling-circle and DNA transposons in the vespertilionid bats. These two TE classes have been largely dormant in most mammals for the past approximately 40 million years and recent insertions are essentially absent from other boreoeutherian genomes31. These results add to previous findings revealing a substantial diversity in TE content within bats, with some species exhibiting recent and ongoing accumulation from TE classes that are extinct in most other mammals while other species show negligible evidence of TE activity32.