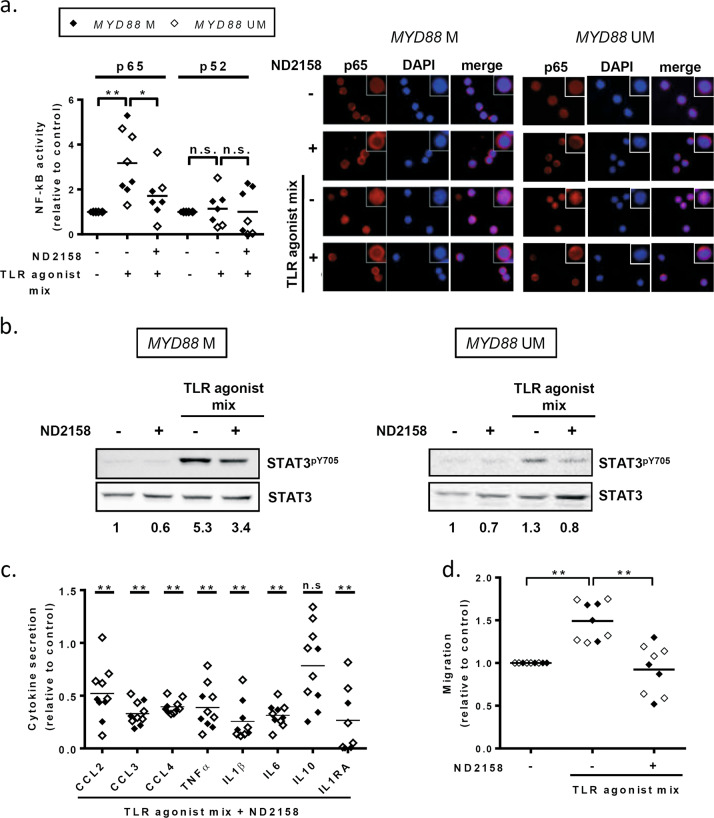
Fig. 4.
Impact of ND2158 on CLL cell signaling, cytokine release and migration. CLL cells were stimulated in vitro with TLR agonist mix for 30 min, before 10 µM ND2158 was added to the culture. a Left panel: Binding of p65 or p52 to NF-κB consensus sequence was analyzed using nuclear extracts from CLL cells of MYD88-mutated (n = 4) and MYD88-unmutated (n = 4) samples via a DNA-binding ELISA-based assay 3 h after treatment. Values are represented relative to untreated samples. Right panel: p65 translocation to the nucleus was analyzed by immunofluorescence microscopy in a representative MYD88-mutated (#7) and MYD88-unmutated (#19) CLL case 3 h after treatment. p65 was stained with anti-NF-κB p65 antibody (clone D14E12) for 30 min, and incubated with an Alexa546-conjugated secondary antibody (red), and DAPI (blue) was used to stain the nuclei. b Western blot analysis for STAT3pY705 of CLL cell extracts of a representative MYD88-mutated (#1) and MYD88-unmutated CLL case (#19) 3 h after treatment. Ratios of phosphorylated and total protein levels were calculated and provided numbers are fold changes relative to the untreated control sample. c Cytokine secretion in supernatants from CLL cells (n = 4 MYD88-mutated; n = 6 MYD88-unmutated) exposed to TLR agonist mix prior treatment with ND2158 for 48 h was analyzed by a multiplexed sandwich immunoassay based on flow cytometry using Luminex® Bead Panel. Values are presented relative to untreated control. Asterisks indicate statistical significance level relative to control. d Migration of TLR-stimulated CLL cells treated with ND2158 for 3 h (n = 4 MYD88-mutated; n = 5 MYD88-unmutated) towards CXCL12 was analyzed by transwell assays. Values are presented as the ratio of migrating cells and total viable cells, relative to the untreated control. Wilcoxon matched-pairs signed-rank test was used for statistical analysis. Horizontal bars represent population means. n.s. not significant; P ≥ 0.05, *P < 0.05, **P < 0.01. M mutated, UM unmutated