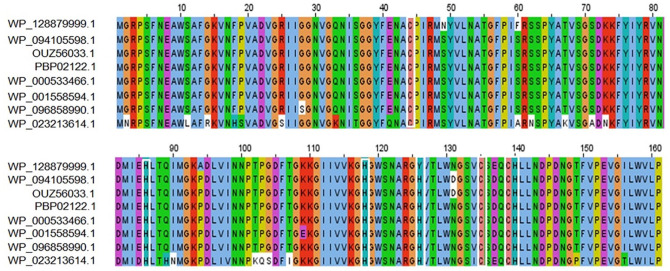
Figure 1.
MSA among different amidase effector 4 (Tae4) proteins using ClustalOmega algorithm by Jalview software. (Top row—target protein, Rows 2 and 3—Shigella flexneri, Row 4—Shigella sonnei, Row 5—Enterobacteriaceae, Rows 6 and 7—Escherichia coli, and Row 8—Salmonella enterica). Marked white boxes indicate conserved catalytic cysteine (C) and histidine (H) residues typical of amidases. MSA indicate multiple sequence alignment.