Figure S3.
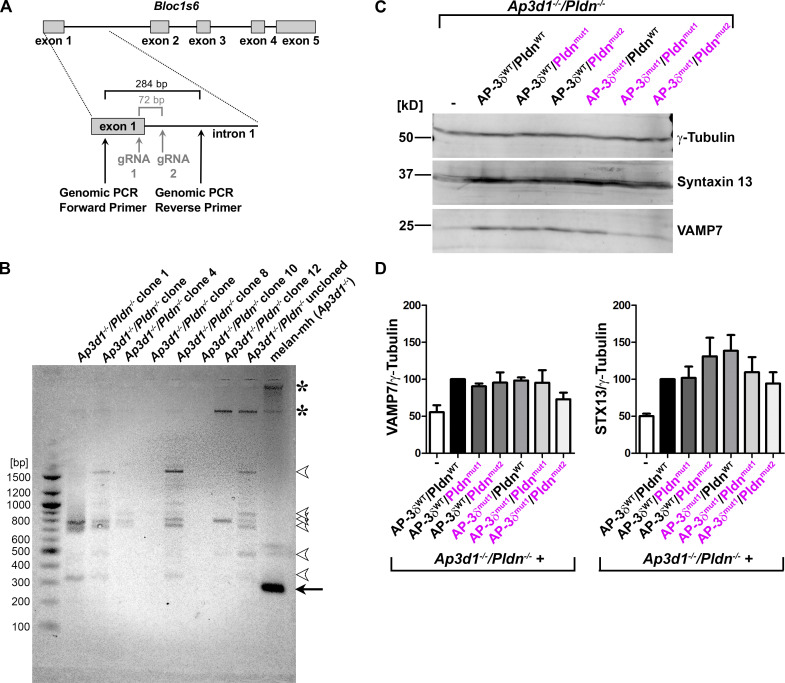
The Bloc1s6 gene is disrupted in Ap3d1−/−/Pldn−/− cells. (A) Schematic diagram of the Bloc1s6 gene (top), encoding Pldn, and the CRISPR/Cas9 mutagenesis and screening strategy for Bloc1s6 gene knockout (bottom) in Ap3d1−/− melan-mh cells. Relative positions of gRNAs 1 and 2 within exon 1 and intron 1 are indicated (gray); if used for precise excision, they would be predicted to remove a 72-bp region as indicated. Positions of genomic primers for PCR in B and the size of the predicted fragment from unmodified Bloc1s6 are indicated. (B) Agarose gel analysis of PCR fragments amplified using the primers indicated in A from genomic DNA of melan-mh, the uncloned population of cells surviving selection following CRISPR/Cas9 mutagenesis (Ap3d1−/−/Pldn−/− uncloned), and 7 of 10 selected Ap3d1−/−/Pldn−/− clones. Positions of DNA size standards (bp) are shown at left. Arrow points to the expected 284-bp fragment detected only in parental melan-mh cells. Arrowheads point to faint bands not observed in melan-mh cells that likely represent off-target PCR products. Asterisks, nonspecific bands detected in melan-mh cells. The additional three clones analyzed had banding patterns similar to those of the clones tested here (not shown). (C) Fractionated whole-cell lysates of Ap3d1−/−/Pldn−/− cells that were untransduced or reconstituted with WT (black) or SNARE binding mutants (magenta) of AP-3δ and Pldn were immunoblotted for VAMP7, STX13, and γ-tubulin (loading control). (D) Quantification of STX13 and VAMP7 protein levels (mean ± SEM), normalized to γ-tubulin levels, over seven (STX13) and four (VAMP7) experiments, respectively. None of the values for either STX13 or VAMP7 content among the different cell lines were statistically different by one-way ANOVA.