Figure 5: Reduction in POM121 in wildtype iPSNs recapitulates C9orf72 mediated alterations in specific Nups and Ran GTPase and increases susceptibility to glutamate induced excitotoxicity.
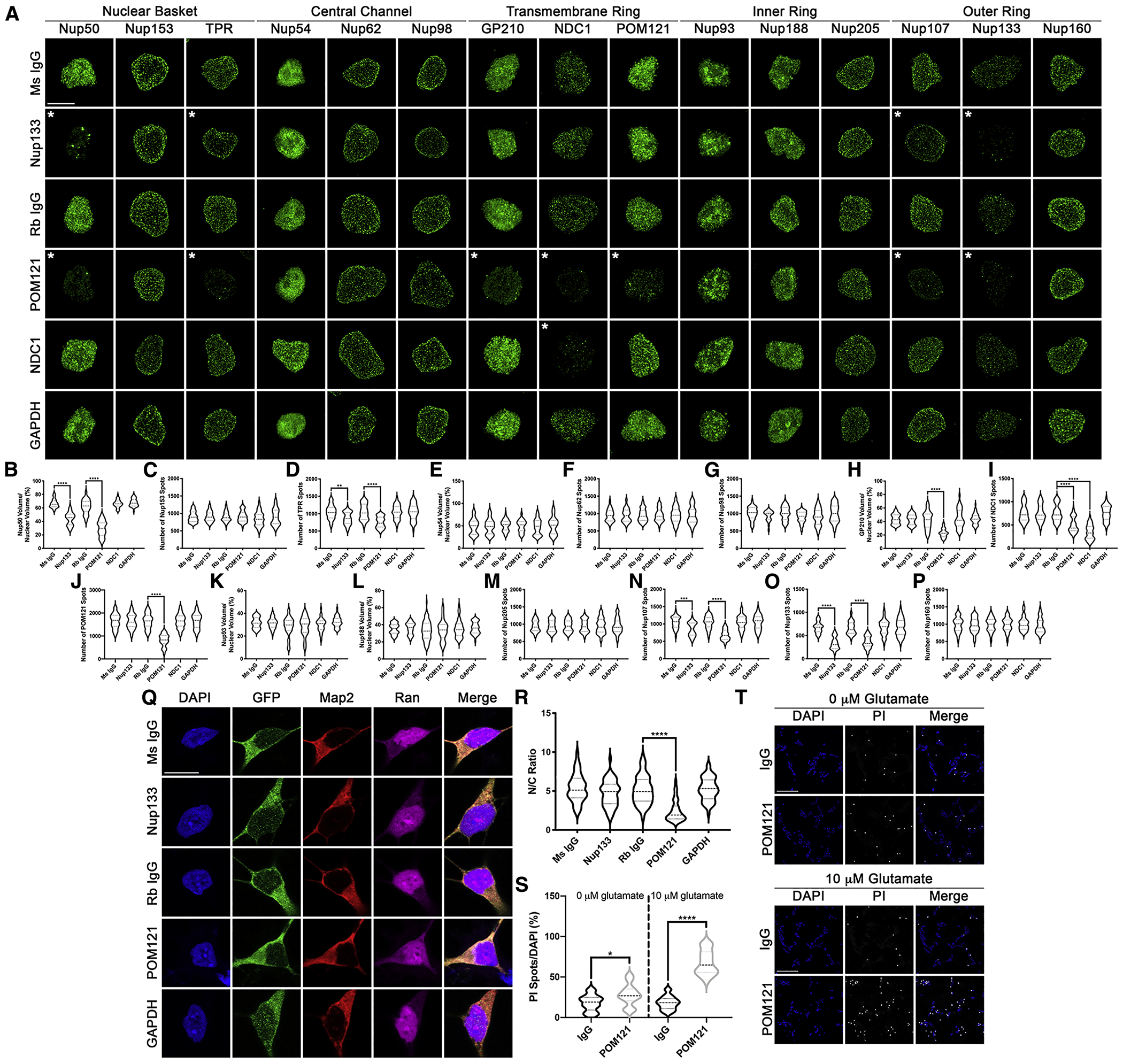
(A-P) Maximum intensity projections from SIM imaging (A) and quantification (B-P) of Nup spots and volume in nuclei isolated from wildtype iPSNs following knockdown of Nup133, POM121, NDC1, or GAPDH. Antibody used for Trim21 GFP mediated knockdown as indicated on left, antibodies as indicated on top. N = 3 wildtype iPSC lines, 50 GFP+ nuclei per line/knockdown. One-way ANOVA with Tukey’s multiple comparison test was used to calculate statistical significance. ** p < 0.01, *** p < 0.001, **** p < 0.0001. (Q) Confocal imaging of control iPSNs following Trim21 GFP knockdown of Nup133, POM121, or GAPDH immunostained for Ran. Antibody used for Trim21 knockdown as indicated on left, antibodies as indicated on top. (R) Quantification of nuclear to cytoplasmic ratio of Ran. N = 3 wildtype iPSC lines, at least 50 cells per line/knockdown. One-way ANOVA with Tukey’s multiple comparison test was used to calculate statistical significance. **** p < 0.0001. (S) Quantification of percent cell death following exposure to glutamate. N = 3 control iPSC lines, 10 frames per well. Two-way ANOVA with Tukey’s multiple comparison test was used to calculate statistical significance. **** p < 0.0001. (T) Confocal imaging of cell death in control and C9orf72 iPSNs as measured by propidium iodide (PI) incorporation. Antibody used for Trim Away as indicated on left, glutamate concentration and stain as indicated on top. Scale bar = 5 μm (A), 10 μm (Q), 100 μm (T). * indicates significantly altered Nups.
