Figure 2.
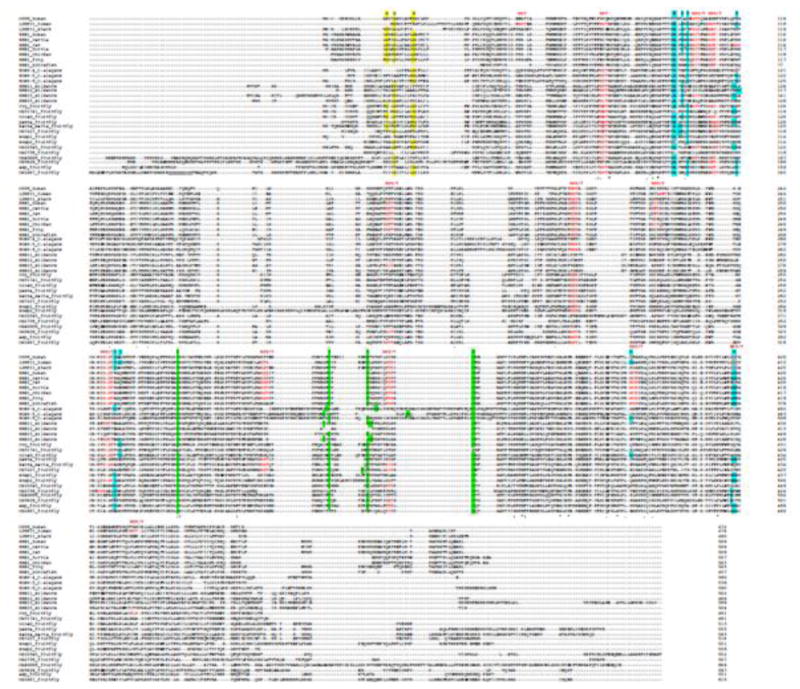
Sequence alignment of SR-BI orthologs from different species. Amino acid sequences from the various SR-BI orthologs were analyzed using the multiple sequence alignment program Clustal Omega from EMBL-EBI (http://www.ebi.ac.uk/Tools/msa/clustalo). Colored amino acids represent the structural features discussed in the text. G_G_G is the glycine dimerization motif, NXS(T) is the N-glycosylation site, C-C is the S-S bond, ERKKDDKE is the tunnel cavity.
