Figure 5.
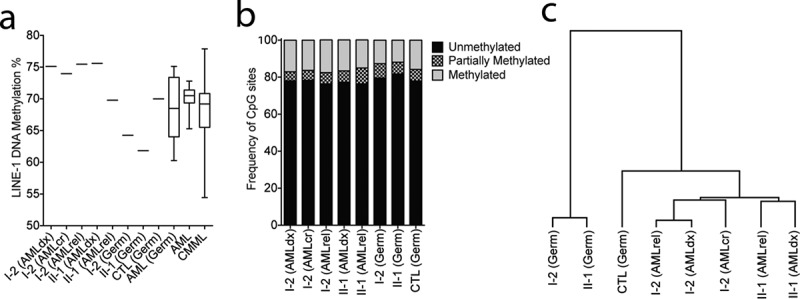
DNMT3A p.P709S is associated with decreased DNA methylation in germline samples
(a) LINE-1 pyrosequencing methylation analysis (PMA) as surrogate of global DNA methylation levels. I-1 (Germ) and II-1 (Germ) have significantly lower methylation than normal oral DNA from a healthy donor (CTL (Germ) and normal oral DNA from 18 AML patients (AML(Germ)) without mutations in any of the following genes: DNMT3A, IDH1, IDH2, TET1, TET2, TET3, DNMT1, DNMT3B, and EZH2 by 295-gene panel sequencing. In comparison, AML samples from I-1 and II-2 present methylation level at and above the upper limits of LINE-1 methylation in primary AML (n = 25) and CMML (n = 69) samples.(b) Genome-wide methylation analysis of germline and AML samples. Reduced Represention Bisulphite Sequencing (RRBS) was performed and there were in total 534,109 CpG sites with a minimum of 20X coverage represented in all samples, excluding sex chromosomes. Frequency of unmethylated (methylation less than 25%), partially methylated (25% to 75% methylation) or methylated (more than 75% methylation) are shown.(c). Hierarchical clustering of the top 1% most variable CpG sites analysed by RRBS. The clustering revealed that patient’s germline samples are most similar, but separated from the other samples.
