Figure 1: A pipeline to generate and analyze standardized morpho-electric data at scale.
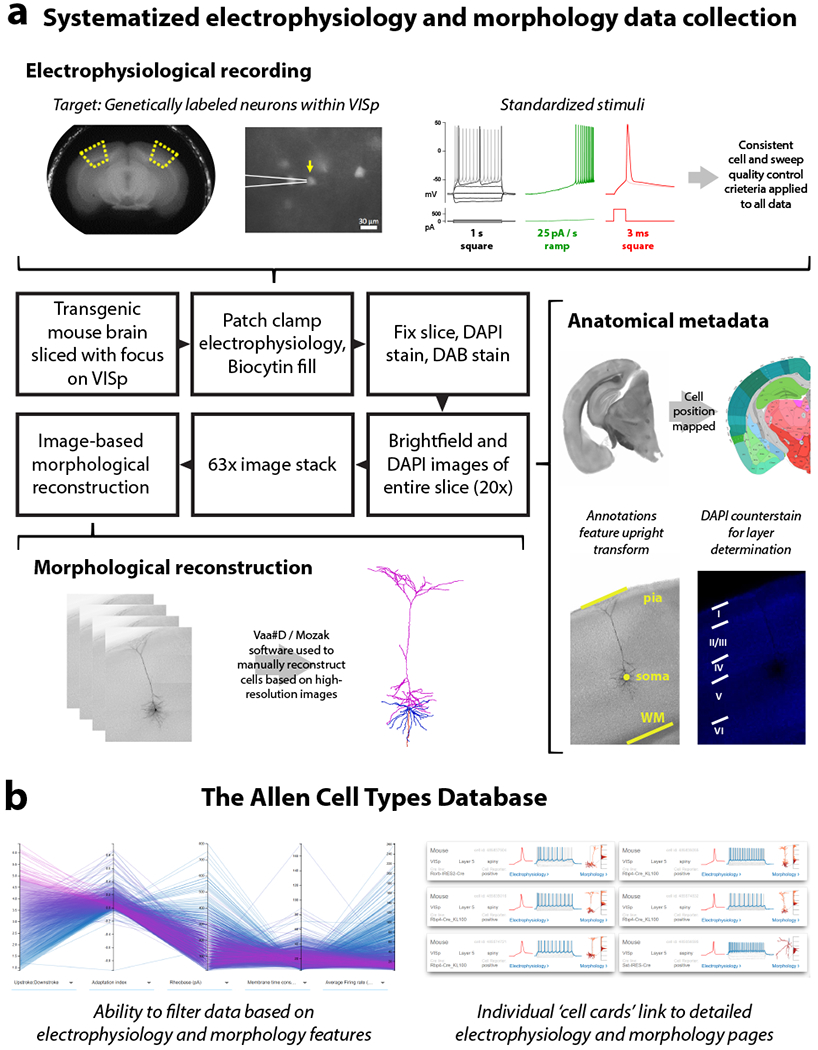
(a) An in vitro single cell characterization pipeline was established to generate standardized electrophysiological and morphological data from mouse cortical neurons. Mouse brains were imaged during vibratome sectioning to aid in cell localization to a common mouse reference atlas, Allen Mouse Common Coordinate Framework version 3 (CCF v3). Fluorescently labeled neurons from specific transgenic mouse lines were recorded by whole cell patch clamping to characterize each cell’s intrinsic electrical properties. Each cell was stimulated with a standard electrophysiological stimulation paradigm and underwent consistent cell and sweep quality control, allowing for routine feature extraction and alignment of data traces from diverse cell types. During the electrophysiology recording, cells were filled with biocytin, then tissue slices were fixed, stained and mounted, and imaged at 20× in a single plane. Cells were mapped to the reference atlas and layer determination was made using a DAPI counterstain. A subset of neurons were then selected to be imaged in a high-resolution 63× stack. High quality cells were then manually reconstructed based on the z-stack images by using the Vaa3D / Mozak software package. (b) Electrophysiology, imaging, and morphology data and metadata for each cell are made freely accessible through the Allen Cell Types Database. An interactive user interface allows users to filter thousands of cells by electrophysiology and morphology features, then each cell has a specific page with detailed electrophysiology and morphology data, when available.
