Figure 1.
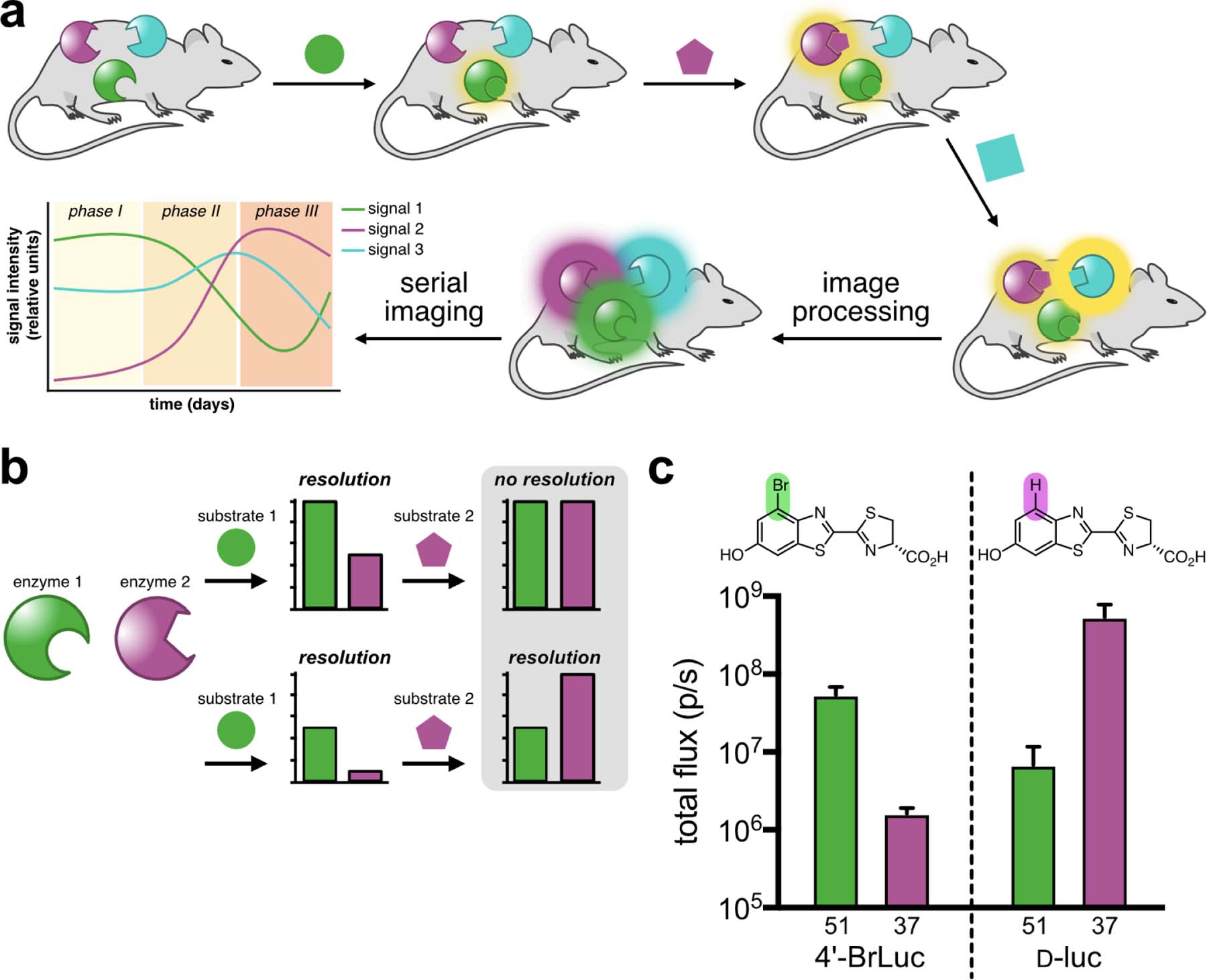
Multicomponent bioluminescence imaging via serial substrate addition and unmixing. (a) Sequential application of orthogonal luciferins (shapes) to illuminate multiple luciferase reporters in vivo. Linear unmixing algorithms can deconvolute substrate signatures, enabling rapid and dynamic readouts of biological processes. (b) Bioluminescent probes must be orthogonal and exhibit differential emission intensities for successful unmixing. When both probes are equally “bright” (top), no resolution is possible. Probes of varying intensity (bottom) can be distinguished when the dimmest probe is administered first. (c) A sample orthogonal bioluminescent pair. Samples expressing mutant 51 or 37 can be resolved using 4’-BrLuc (100 μM) or d-luc (100 μM). Error bars represent the standard error of the mean for n = 3 experiments.
