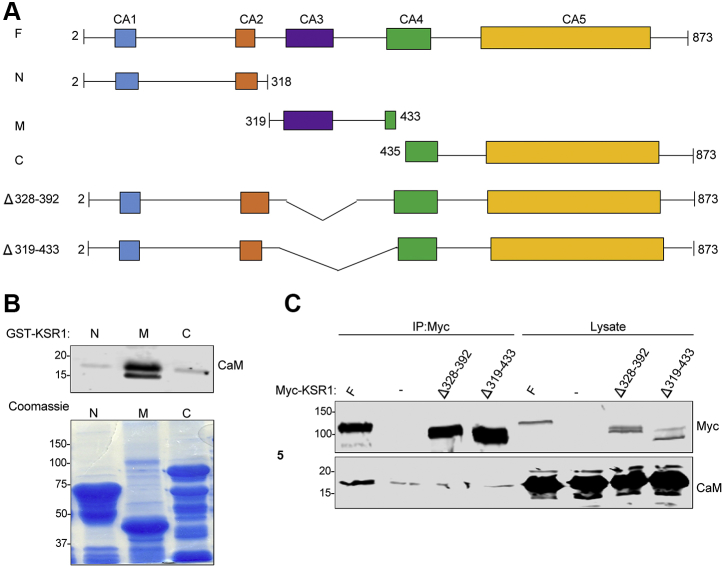
Figure 3.
Identification of the calmodulin-binding region in KSR1.A, schematic representation of KSR1 constructs. KSR1 contains five distinct domains, termed conserved area 1 (CA1) through CA5. The KSR1 constructs are full-length (F, amino acids 2–873), N (2–318), M (319–433), and C (435–873). KSR1Δ328–392 has amino acids 328–392 deleted and KSR1Δ319–433 has amino acids 319 to 433 deleted. B, GST-tagged fragments of KSR1-N, -M or -C were incubated with purified calmodulin. Complexes were pulled down with glutathione-Sepharose beads and analyzed by SDS-PAGE. The gel was cut at ∼25 kDa. The lower portion of the gel was processed by western blotting and probed with anti-calmodulin (CaM) antibody. The upper portion of the gel was stained with Coomassie blue. Data are representative of two independent experiments. C, HEK293 cells were transfected with Myc-tagged KSR1 constructs (F, Δ328–392 or Δ319–433) or not transfected (−). Cells were lysed in buffer containing Ca2+ and complexes were immunoprecipitated (IP) with anti-Myc Affinity Gel. Samples were resolved by western blotting and probed with anti-Myc and anti-calmodulin antibodies. Lysates not subjected to immunoprecipitation were processed in parallel (Lysate). Data are representative of two independent experiments. The positions of migration of molecular mass markers are indicated on the left of the blots and gels.