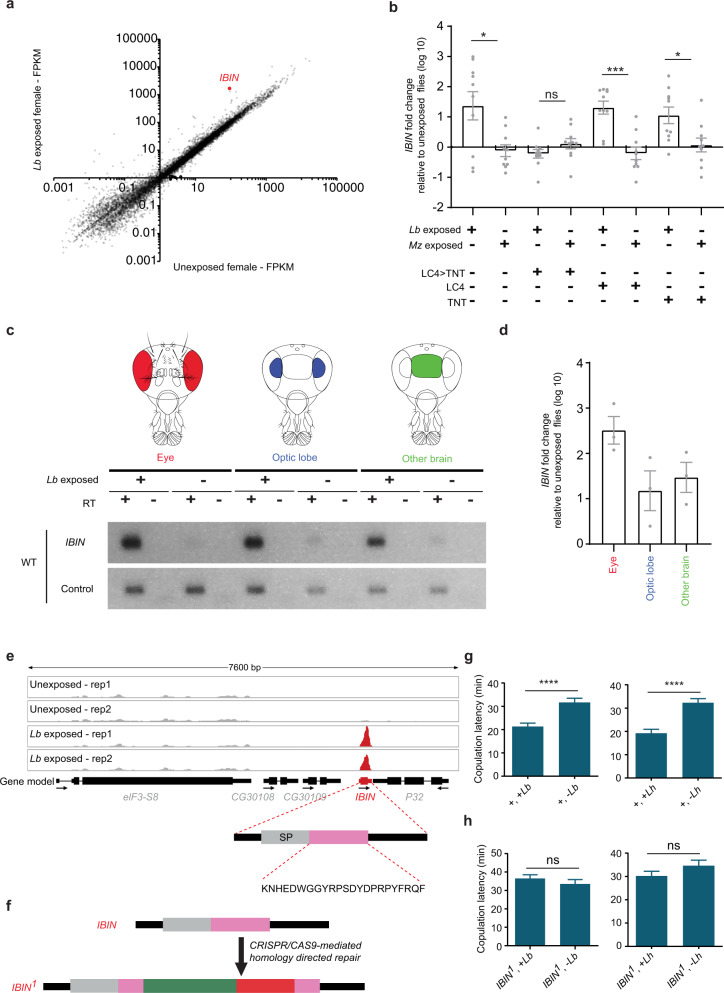
Fig. 5. Exposure to wasps affects gene expression.
a Head transcriptomes of flies that have been in visual contact with L. boulardi for 2 h, and of control flies. The transcriptome is based on two biological replicates. FPKM fragments per kilobase of transcript per million mapped reads. b RT-qPCR analysis of IBIN. Exposure to L. boulardi or M. zaraptor was for 2 h. p = 0.046, 0.07, 0.0008, 0.019 (from left to right), **p < 0.01, Mann–Whitney U test, two-tailed, n = 10. Error bars = SEM. c RT-PCR amplification from the indicated tissues. The eye tissue includes both retina and lamina. RT reverse transcriptase. The control gene is eIF3c. RT(−) refers to a negative control where no reverse transcriptase is added to ensure that genomic DNA is not being amplified. n = 2. d RT-qPCR analysis of IBIN from the indicated dissected tissues of flies that had been in visual contact with L. boulardi for 2 hv. from unexposed control flies. n = 3. Error bars = SEM. e RNA-seq reads (y-axis = 0–8000 reads), n = 2. SignalP (version 5.0) was used to predict the presence of signaling peptide (SP in figure) and the cleavage site, and DeepLoc (version 1.0) was used to predict its extracellular localization. f Generation of a mutant IBIN1 allele. Green indicates core promoter GAL4 sequences; red indicates 3XP 3-DsRed sequences. g, h Copulation latency of IBIN1 mutant flies and wCS control flies that had been paired with wasps (L. boulardi). Two-tailed Mann–Whitney test, n = 67 in all cases. Error bars = SEM. In g, p < 0.0001 (left and right). In h, p = 0.377 (left), p = 0.1718 (right).