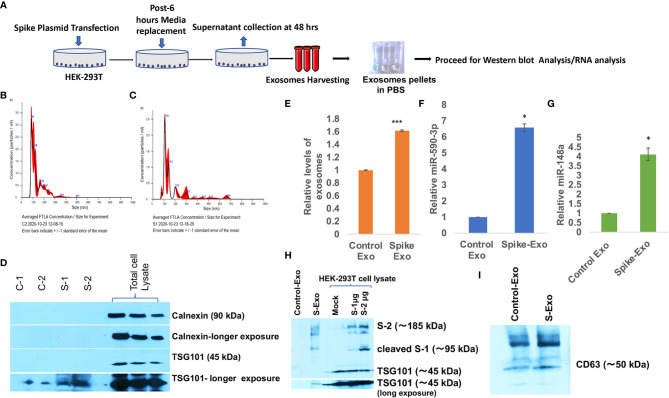
Figure 1.
Spike transfected cells released exosomes are loaded with miR-148a and miR-590. (A) Work-flow schematic depicting exosome harvesting protocol from Spike gene transfected HEK-293T cells. (B) Size distribution analysis graph of control exosomes done on NanoSight, NTA 3.2 Dev Build 3.2.16, indicating mode of size range 90.6 nm. (C) Size distribution analysis graph of S-exosomes, indicating mode of size range of 107.5 nm. (D) Immunoblot image for characterization of harvested exosome preparations. Endosomal protein Calnexin is absent in exosome population indicating the purity of exosome and free of cellular debris. TSG101 as tetraspanin marker protein are present in exosomes. (E) Graph bars showing relatively higher amount of exosome secretion by Spike transfected cells, measured as total protein via Bradford assay. (F) Graph bars displaying relative miR-590 levels in Spike-Exosomes compared to control exosomes. (G) Graph bars displaying relative levels of miR-148a in Spike-Exosomes as compared to control exosomes. Exosomes pellets were subjected to total RNA isolation and microRNA levels measured through qPCR analysis via TaqMan assay specific for miR-590 and miR-148a respectively. (H) Western blot image, confirming the Spike gene transfection in HEK-293T cells as well their accumulation in secreted exosomes. 1µg and 2 µg of Spike gene (S) plasmids (pTwist-EF1α-nCoV-2019-S-2×Strep) were transfected in 6-well plate format for 24 hours with the help of Lipofectamine 2000. (I) Western blot image showing CD63 in harvested exosomes as exosomal marker. All the experiments have been repeated at least three times independently to obtain mean and mean ± S.E.M. Levels of significance have been calculated by student’s t-test and statistical significance indicated as single * for p values <0.05 and *** for p values <0.0005.