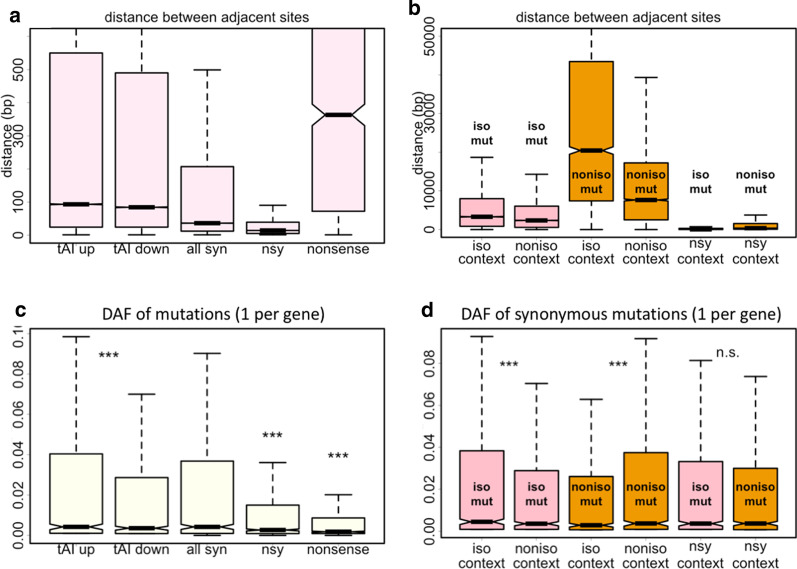
Fig. 3.
Control for distance to cancel the effect of LD. a Distance (bp) between adjacent mutations sites. Mutations were classified into tAI up, tAI down, synonymous, nonsynonymous, and nonsense. Each type of mutations was calculated separately. b Distance (bp) between adjacent mutations sites. Mutations were classified according to their context. Each type of mutations was calculated separately. c DAF of CDS mutations of different categories. Mutations were classified into tAI up, tAI down, synonymous, nonsynonymous, and nonsense. We only selected one mutation per gene (the most 5-prime one) to exclude the effect of LD. d DAF of CDS mutations of different categories. Mutations were classified according to their context. We only selected one mutation per gene (the most 5-prime one) to exclude the effect of LD. Wilcoxon rank sum test was used to calculate p-value. ***Represents p-value < 0.001; n.s., not significant. Multiple testing correction was performed to adjust the p-value. Length of multiple testing correction was set to the number of tests performed