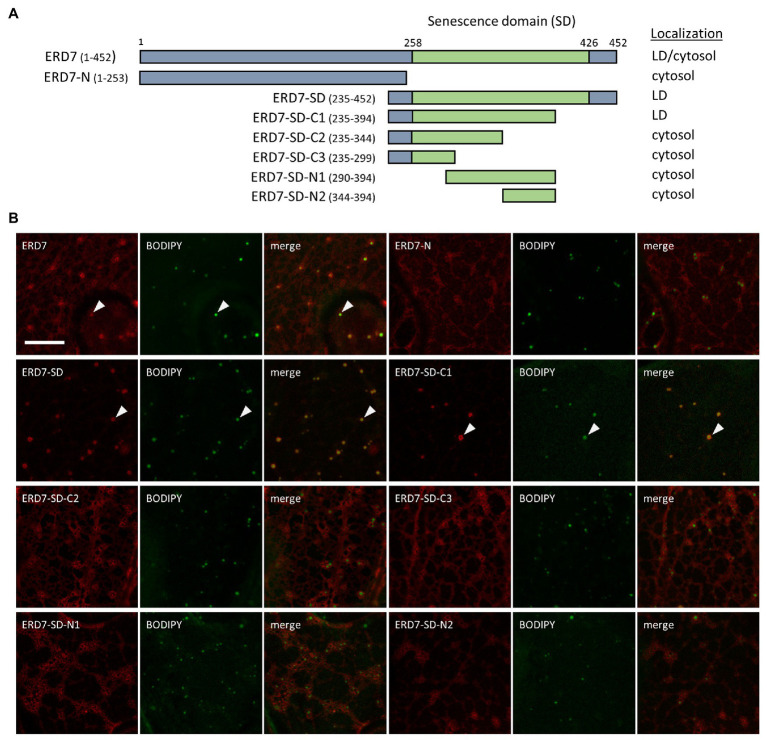
Figure 2.
Subcellular localization of various ERD7 truncation mutants in N. benthamiana leaves. N. benthamiana leaves were transiently transformed (via Agrobacterium infiltration) with either full-length or truncated versions of ERD7 and tagged at their N-termini with mCherry. Three days post-infiltration, leaves were stained with the neutral lipid dye BODIPY and imaged using CLSM. Shown in (A) are schematic representations of full-length ERD7 and the various ERD7 mutants and their corresponding subcellular localization(s) [i.e., cytosol and/or lipid droplet (LD)]. Numbers above the illustration of full-length ERD7 represent specific amino acids residues, and the SD (amino acid residues 258–426) is defined based on information obtained from the InterPro database. The numbers next to the name of each construct denote the amino acid residues in ERD7 that were fused to mCherry; note that the N-terminal-appended mCherry moiety is not depicted in the illustrations or construct names. The portion of ERD7 representing the SD or the other parts of the protein are depicted in the illustrations as green or gray boxes, respectively. Shown in (B) are representative CLSM images (z-sections) of each mCherry-ERD7 (mutant) protein, as indicated by the panel labels, along with the corresponding BODIPY-stained LDs in the same cell. Also shown is the corresponding merged image. Arrowheads indicate examples of mCherry-ERD7 and the selected (mutant) versions thereof (i.e., ERD7-SD and ERD7-C1) that localized to BODIPY-stained LDs; compare with the lack of LD localization for the other mutant versions of ERD7. Scale bar in (B) = 10 μm.