Figure 4.
Assay validation and estimation of true intactness
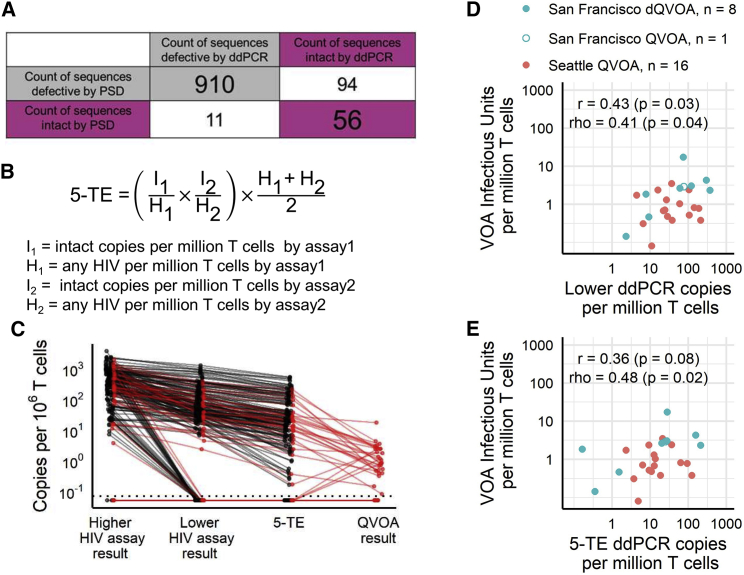
(A) 2 × 2 table showing the number of sequences with predicted intact or defective calls using our protocol and the calls based on sequence analysis in the Proviral Sequence Database.
(B) Equation to calculate the expected number of HIV genome copies per 106 T cells that would give triple-positive results from both HIV assays (5-target estimate [“5-TE”]): the probability of intactness by assay1 and intactness by assay2, multiplied by the average number of HIV copies (both intact and defective) that were detected across the two assays.
(C) Approximation of true provirus intactness using the ddPCR protocol and relationship to quantitative viral outgrowth assay (QVOA). Shown are the lower and higher of the two HIV assay results (both n = 192), the 5-TE n = 192), and the QVOA result where available (red lines; n = 35).
(D and E) Correlation between QVOA or dQVOA and the lower of the two assays (D) or 5-TE (E). Not included are samples where QVOA or ddPCR results were zero. Spearman’s rho and Pearson’s r are given, with p values resulting from tests of the null hypotheses that there is no monotonic (Spearman) or linear (Pearson) relationship between the two parameters. For the UW-CFAR_QVOA cohort (n = 14 PLH), two samples were tested at two different dilutions. For the San Francisco cohort (n = 9 PLH), one sample did not have a dQVOA result but did have a QVOA result (open blue circle). All tests were done with three replicate wells for the HIV assays and two replicate wells for the reference gene assay.