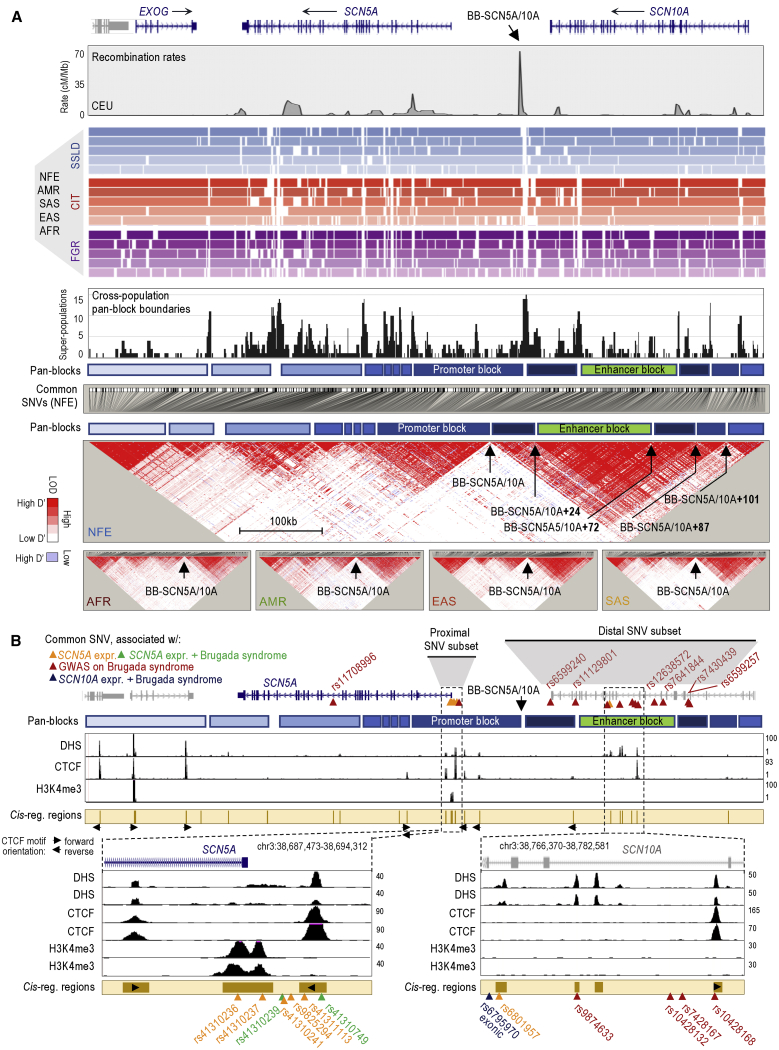
Figure 1.
Haplotype block frameworks in the SCN5A-SCN10A locus
(A) The top profile shows HapMap rate estimates of recombination in the CEU population in cM (centimorgans)/Mb (megabase). Genomic coordinates (hg19): chr3:38,516,506-38,841,720. The colored tracks depict haplotype-block estimates by super-population (from top to bottom: NFE, AFR, AMR, EAS, and SAS) using 3 block-partitioning methods (SSLD, CIT, and FGR) implemented in Haploview version 4.2. A summary profile of the predicted pan-blocks/pan-boundaries (in black) shows the sum of the times a block boundary is imputed in a population with a given method (max value = 15 for 5 populations with three methods). Note: <100-bp boundaries may not be visible in the colored tracks, yet will be accounted for in the summary profile. Tracks in blue represent the approximated locations of the predicted pan-blocks/pan-boundaries (top track: relative to genomic coordinate; bottom track: relative to SNV). The large heatmap represents a LD plot of n = 697 SNVs with MAF ≥5% in NFE, and the spatial distribution of LD coefficient (D’) values and the likelihood of odds (LOD) ratios for each pairwise SNV set based on the 1KG Phase 3 dataset. The rest of LD plots correspond to n = 967 AFR, n = 838 AMR, n = 728 EAS, and n = 736 SAS. Predicted pan-boundaries in the NFE plot are indicated: BB-SCN5A/10A (chr3:38,724,850-38,727,325), BB-SCN5A/10A+24 (chr3:38,751,191-38,752,018), BB-SCN5A/10A+72 (chr3:38,798,839-38,799,304), BB-SCN5A/10A+87 (chr3:38,814,174-38,814,220), and BB-SCN5A/10A+101 (chr3:38,828,333-38,829,269). Only the BB-SCN5A/10A site is indicated in the rest of the LD plots. See also Data S1, Table S1.
(B) Genomic location of relevant common SNVs and maps of DHS, CTCF occupation, and H3K4me3 accumulation in human cardiac myocytes (data sources: GSM736516/GSM736504, GSM1022657/GSM1022677, and GSM945308). The cantaloupe-colored track depicts the sum of chromatin features. CTCF motifs and orientation also indicated. Genomic coordinates based on hg19. See also Figure S1.