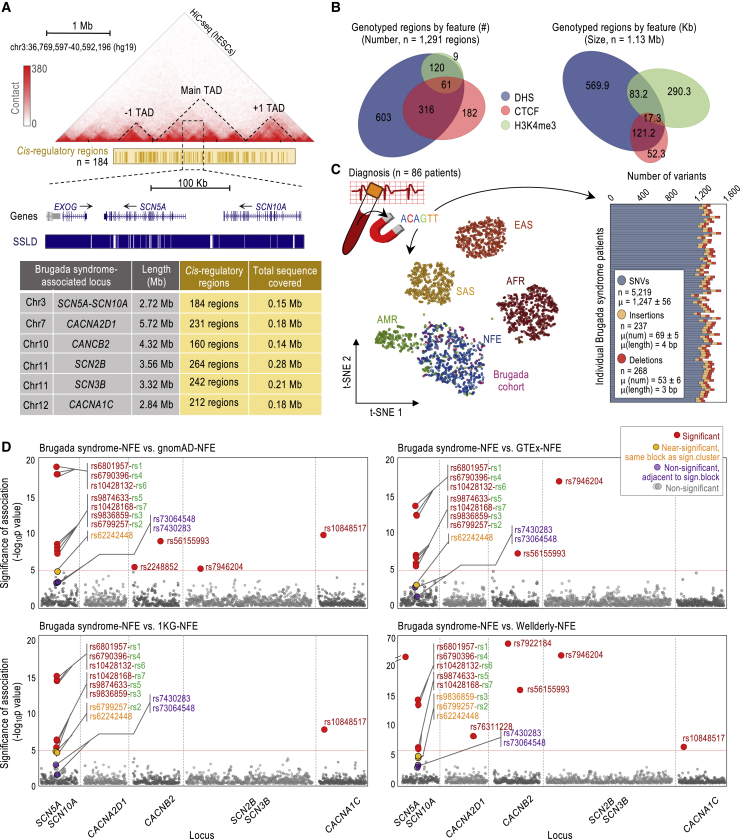
Figure 2.
Deep genotyping of cis-regulatory regions in Brugada-associated loci
(A) Heatmap of long-range chromatin interactions to delineate TAD structure (Hi-C-seq; data source: GSM862723/GSM892306). TAD boundaries set according to Dixon et al.47 Cantaloupe-colored track depicts DHS/CTCF/H3K4me3 regions across 3 TADs in human cardiac myocytes (data sources: GSM736516/GSM736504, GSM1022657/GSM1022677, and GSM945308). Blue tracks depict SSLD-based haplotype-block estimations. Table shows the size (Mb) of the 3 TADs selected for each Brugada syndrome-associated locus, the number of cis-regulatory regions, and their total size (Mb).
(B) Summary of cis-regulatory regions by chromatin feature (by class or base pairs). The 2 manually included regions are represented only on the right panel. See also Data S1, Table S2.
(C) Ancestry and variant annotation based on genotyping of cis-regulatory regions captured from blood-derived genomic DNA of n = 86 Brugada syndrome cases (type I ECG-based diagnosis). The t-SNE plot, based on the first 6 principal components, shows the ancestry admixture for the Brugada syndrome cases in the context of the 1KG Phase 3 human super-populations. Graph shows the summary of SNVs/insertions/deletions in the Brugada cohort (total number, n; mean number, μ [number]; and mean size μ [length] for indels). See also Figure S2 and Data S1, Table S3.
(D) Manhattan plots showing the significance of the associations for n = 2,121 common SNVs in the Brugada cohort in a case-control analysis of n = 86 Brugada syndrome cases using n = 7,718 NFE individuals (gnomAD) as controls (top left), n = 355 (GTEx; top right), n = 404 (1KG Phase 3; bottom left), and n = 196 (Wellderly; bottom right) NFE individuals. Relevant SNVs labeled as indicated. Significance tested by Fisher’s exact test. The red line marks the threshold of significance (Bonferroni-corrected α level of p value 2.36e−5 [0.05/2,121] according to the number of common SNVs tested). See also Figure S2 and Data S1, Table S4.