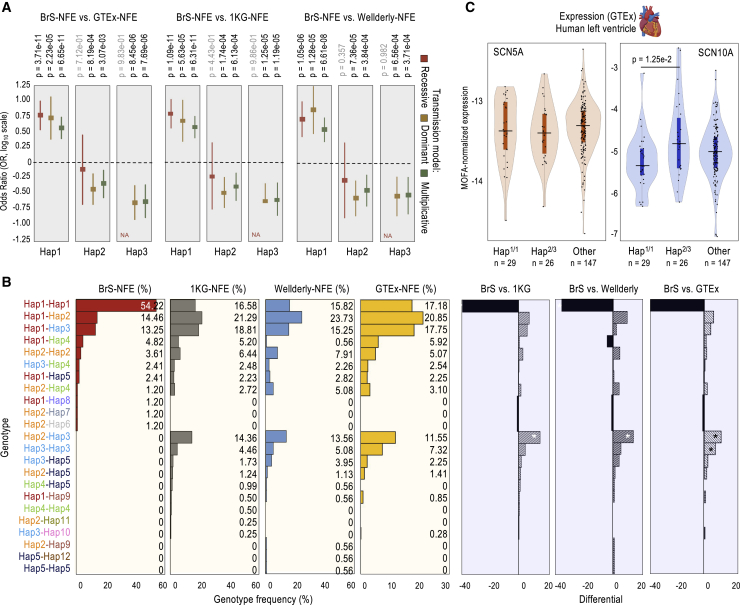
Figure 4.
Transmission models, genotypes, and functional associations
(A) Forest plot showing OR estimates and 95% confidence intervals (CIs) for the association between carrying the Hap1, Hap2, or Hap3 variants and risk of Brugada syndrome (BrS). ORs were obtained using logistic regression models assuming a recessive, dominant, or multiplicative inheritance after adjusting for gender. Horizontal lines indicate OR = 1. OR > 1 is associated with high risk of Brugada syndrome; OR < 1 is associated with lower risk of Brugada syndrome. Bonferroni-corrected p values indicated; significance threshold p < 3.33e−3, based on the number of total haplotypes tested (0.05/15). See also Data S1, Table S6.
(B) Frequency of genotypes in the Brugada-NFE (n = 83), 1KG-NFE (n = 404), Wellderly-NFE (n = 177), and GTEx-NFE (n = 355) cohorts/datasets (percentages indicated), and differential frequency for the indicated comparisons. Significance tested by Fisher’s exact test. Results were considered significant (∗) if p values were below a Bonferroni-corrected α level of 1.79e−3 (0.05/23) based on the number of genotypes tested. See also Data S1, Table S6.
(C) cis-eQTL analysis of the Hap1/1, Hap2/3, and the rest of genotypes (other), as in (B), using expression data of human left ventricle tissue generated by GTEx (no ancestry selection; MOFA-corrected expression). Violin plot shows median expression and box indicating interquartile range and sample point (number also indicated). Significance tested by 1-way ANOVA and Tukey honest significant differences (Tukey HSD) test for multiple pairwise comparisons. Significance threshold, p < 0.05. Significant case indicated. Rare genotypes only observed in the GTEx dataset (Hap2/14, Hap3/14, Hap3/9, Hap1/13, and Hap1/15) are not included. See also Figure S4.