Summary
Genetic manipulation of neural precursor cells is an important tool to study mechanisms underlying proliferation, fate specification, and neuron formation. The CRISPR/Cas9 system enables efficient genome editing but requires the clonal expansion of cells containing the desired mutation. Here, we describe a protocol for the effective generation of clonal mouse hippocampal neural precursor lines with CRISPR/Cas9-based gene knockouts. Edited cell lines can be used to investigate gene regulatory networks driving neuronal differentiation and for modeling of diseases that involve hippocampal neurogenesis.
For complete details on the use and execution of this protocol, please refer to Pötzsch et al. (2021).
Subject areas: Cell biology, Cell-based Assays, Genetics, Molecular biology, CRISPR, Neuroscience, Stem cells
Graphical abstract

Highlights
-
•
Generation of clonal mouse hippocampal neural precursor cell lines
-
•
Step-by-step protocol for producing CRISPR/Cas9-based gene knockouts
-
•
Description of single guide RNA and dual guide RNA knockout approaches
-
•
Validation of gene knockouts by PCR and sequencing and representative analyses
Genetic manipulation of neural precursor cells is an important tool to study mechanisms underlying proliferation, fate specification, and neuron formation. The CRISPR/Cas9 system enables efficient genome editing but requires the clonal expansion of cells containing the desired mutation. Here, we describe a protocol for the effective generation of clonal mouse hippocampal neural precursor lines with CRISPR/Cas9-based gene knockouts. Edited cell lines can be used to investigate gene regulatory networks driving neuronal differentiation and for modeling of diseases that involve hippocampal neurogenesis.
Before you begin
This protocol describes the generation of gene knockouts in neural precursor cells derived from the adult mouse hippocampus. We outlined the specific steps for producing indels (small insertions or deletions) and for deletions of larger gene regions through CRISPR/Cas9-mediated non-homologous end joining.
Detailed protocols on the isolation of neural precursor cells from the adult mouse hippocampus have been published elsewhere (Babu et al., 2011; Walker and Kempermann, 2014; Bernas et al., 2017). For alternative protocols describing gene editing of mouse and human neural precursor cells please refer to (Bressan et al., 2017; Dewari et al., 2018; Dever et al., 2019).
Before start of the protocol, neural precursor cell cultures should be prepared and oligonucleotides for cloning of sgRNAs designed and ordered.
Culture conditions for hippocampal neural precursor cells
Timing: 3 days
-
1.Cell culture dishes: neural precursor cells are cultured on poly-D-lysine/laminin-coated surfaces.
-
a.For every gene knockout cell line you wish to generate, coat 2× T25-flasks, 1× 96-well-plate, 4× 6-well-plates. Coating protocol:
-
i.Add poly-D-lysine (5 μg/mL in ddH2O) to dishes and incubate for 6 h at 20°C–23°C (volume of poly-D-lysine solution: 3 mL/T25 flask; 100 μL/96-well; 1 mL/6-well).
-
ii.Wash dishes three times with ddH2O and dry surfaces for 10 min at 20°C–23°C.
-
iii.Add laminin (5 μg/mL in cold DMEM/F-12) to dishes and incubate for 10–16 h at 37°C in a cell culture incubator (volume of laminin solution: 3 mL/T25 flask; 100 μL/96-well; 1 mL/6-well).
-
iv.Store dishes with laminin solution at −20°C until usage. Coated dishes can be stored for up to 3 months.
-
i.
-
a.
-
2.Cell culture medium: neural precursor cells are cultured in proliferation medium with half-medium changes every two days.
-
a.For the recipe of proliferation medium please see the materials and equipment section.
-
b.Incubate cells in a cell culture incubator.
-
c.Half medium change: Remove 50% of medium from the flask and replace it with fresh proliferation medium.
-
a.
-
3.Passaging of cells: neural precursor cells are passaged at a confluence of 80%. Neural precursor cells can be used for experiments for up to 20 passages after isolation from the hippocampus.
-
a.Remove culture medium and wash once with PBS.
-
b.Incubate cells with Accutase for 5 min at 37°C. Add 0.5 mL of Accutase per T25 flask (20 μL/cm2).
-
c.Resuspend cells in wash medium (10× volume of Accutase), transfer to 15 mL tube and spin for 5 min at 300 × g.
-
d.Remove supernatant, resuspend cell pellet in fresh proliferation medium and plate on dishes with a seeding density of 20,000 cells/cm2.
-
a.
Design of sgRNAs and genotyping primers
Timing: 1 day
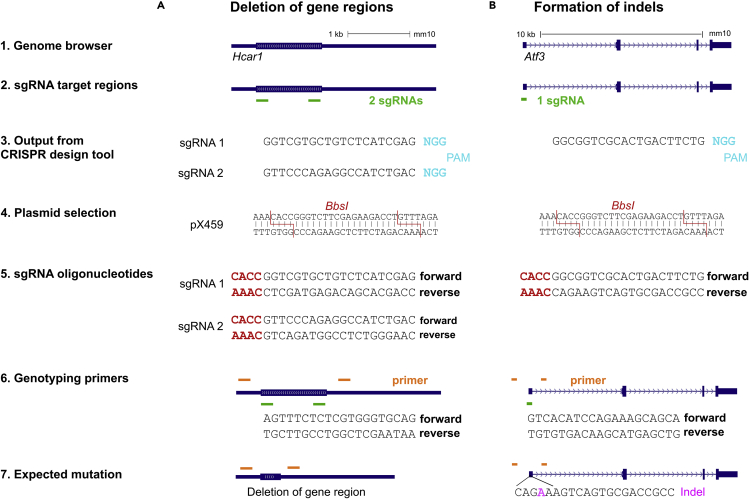
This section provides a brief description of how to design sgRNAs for CRISPR/Cas9-based gene knockouts (Figure 1). More detailed considerations for sgRNA design have been published elsewhere (Ran et al., 2013; Cui et al., 2018).
-
4.
Retrieve the sequence of the mouse target gene from a genome browser such as the Ensembl Genome Browser (http://www.ensembl.org/Mus_musculus/Info/Index).
-
5.
Design sgRNAs targeting the gene of interest using a CRISPR design tool such as E-CRISP (http://www.e-crisp.org/E-CRISP/). For generating small frameshift mutations (indels), design one sgRNA. For deletion of gene regions, design two sgRNAs flanking the desired region.
Note: Knockouts of protein coding genes can be generated through both the formation of indels (single sgRNA approach) or the deletion of gene regions (dual sgRNA approach). Advantages of the dual sgRNA approach include 1) that specific regions of a gene can be knocked-out (also those located at the 5′ end of a gene) and 2) that they can be quickly and easily detected by PCR. Moreover, the dual sgRNA approach can also be used for perturbing non-coding regions or genes encoding microRNAs or long non-coding RNAs. The advantages of the single sgRNA approach lie in the high efficiency of indel formation and that only one sgRNA is needed, which reduces the risk for potential off-target activities and the work-load during cloning. Moreover, the single sgRNA approach can be used when the sequence of a specific target gene does not enable the design of two sgRNAs.
Note: The advantage of CRISPR design tools is that they calculate and report probabilities for off-target activity for each sgRNA. Prioritize sgRNAs with a high specificity score (S-score in E-CRISP).
Alternatives: An overview of CRISPR design tools other than E-CRISP can be found at https://zlab.bio/guide-design-resources.
-
6.
Retrieve the respective sgRNA nucleotide sequence from the CRISPR design tool.
CRITICAL: The sgRNA target sequence has to precede a protospacer adjacent motif (PAM) sequence in the genome, which is necessary for the endonuclease activity of Cas9. For instance, Cas9 from Streptococcus pyogenes requires that sgRNA target sequences are followed by 5′-NGG-3′ (with “N” being any nucleotide). Although the PAM sequence is depicted in the output sequence of E-CRISP and most other design tools, it should NOT be included in the sgRNA sequence.
-
7.
Generate the reverse complement of the sgRNA sequence.
Note: While the output from E-CRISP will be the scaffold of the forward sgRNA oligonucleotide, its reverse complement will yield the scaffold of the reverse sgRNA oligonucleotide. Forward and reverse sgRNA oligonucleotides will later be hybridized to enable sgRNA cloning.
CRITICAL: For efficient transcription, sgRNAs should start with a guanidine residue at their 5′ end. E-CRISP has an in-built filter for this criterion, however, if you use other design tools, make sure that forward oligonucleotides start with a guanidine residue or add an extra guanidine (and a cytidine residue at the reverse oligonucleotide).
-
8.
Add the flanking nucleotides to the sgRNA oligonucleotides, which are required for insertion into the plasmid. For cloning into the BbsI site of vector pX459, add CACC at 5′ of the forward oligonucleotide and AAAC at 5′ of the reverse oligonucleotide.
Alternatives: A variety of other plasmids that enable CRISPR/Cas9-based gene editing is available from Addgene (https://www.addgene.org/crispr).
-
9.
Design primer pairs flanking the sgRNA target site for polymerase chain reaction (PCR). Those primers will be used for genotyping of the cell lines and validation of gene knockouts.
-
10.
Design sgRNA oligonucleotide against lacZ, which will be used to generate a control cell line. Potential sequence for lacZ forward sgRNA oligonucleotide: GTGCGAATACGCCCACGCGAT.
-
11.
Order oligonucleotides from a commercial provider in desalted form and at minimal scale.
Figure 1.
Design of sgRNAs for generating gene knockouts
(A) Dual sgRNA approach for generating gene knockouts by deletion of gene regions. As an example, the gene Hcar1 is illustrated, which has a coding sequence length of 1056 bp located on a single exon. Depicted is a strategy for the knockout of Hcar1 through deletion of a 974 bp sequence from its exon.
(B) Single sgRNA approach for generating knockouts through indel formation. Depicted is the gene Atf3, which has a length of 10 kb. To induce a frameshift mutation in Atf3, a sgRNA was designed to target the first coding exon, which is common to all known splice variants.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| One ShotTM TOP10 Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C404010 |
| Chemicals, peptides, and recombinant proteins | ||
| GlutaMAXTM Supplement | Thermo Fisher Scientific | Cat# 35050061 |
| B-27TM Supplement | Thermo Fisher Scientific | Cat# 17504044 |
| Penicillin-streptomycin | Thermo Fisher Scientific | Cat# 15140122 |
| Human EGF | PeproTech | Cat# AF-100-15 |
| Human FGF-2 | PeproTech | Cat# 100-18B |
| Heparin | MP Biomedicals | Cat# 0210193125 |
| Poly-D-lysine | Sigma-Aldrich | Cat# P7280 |
| Laminin | Roche Diagnostics | Cat# 11243217001 |
| T4 DNA Ligase | Thermo Fisher Scientific | Cat# EL0014 |
| FastDigest BbsI | Thermo Fisher Scientific | Cat# ER1011 |
| T4 Polynucleotide Kinase | Thermo Fisher Scientific | Cat# EK0031 |
| StemProTM AccutaseTM | Thermo Fisher Scientific | Cat# A1110501 |
| Puromycin-Dihydrochloride | Thermo Fisher Scientific | Cat# A1113803 |
| Critical commercial assays | ||
| LipofectamineTM 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L30001 |
| GeneJET Plasmid Miniprep Kit | Thermo Fisher Scientific | Cat# K0503 |
| GeneJET Endo-Free Plasmid Maxiprep Kit | Thermo Fisher Scientific | Cat# K0861 |
| DMEM-F12 without glutamine | Thermo Fisher Scientific | Cat# 21331020 |
| NeurobasalTM medium | Thermo Fisher Scientific | Cat# 21103049 |
| Phusion High-Fidelity DNA Polymerase | Thermo Fisher Scientific | Cat# F530S |
| Opti-MEMTM Reduced Serum Medium | Thermo Fisher Scientific | Cat# 31985062 |
| QIAamp DNA Micro Kit | QIAGEN | Cat# 56304 |
| QuickExtractTM DNA Extraction Solution | Lucigen | Cat# QE09050 |
| Experimental models: Cell lines | ||
| Neural precursor cells from adult mouse hippocampus | Laboratory of Gerd Kempermann | Line MI6 |
| Oligonucleotides | ||
| Hcar1_sgRNA1_fw: CACCGGTCGTGCTGTCTCA TCGAG |
(Pötzsch et al., 2021) | N/A |
| Hcar1_sgRNA1_rev: AAACCTCGATGAGACAGCA CGACC |
(Pötzsch et al., 2021) | N/A |
| Hcar1_sgRNA2_fw: CACCGTTCCCAGAGGCCATCTGAC |
(Pötzsch et al., 2021) | N/A |
| Hcar1_sgRNA2_rev: AAACGTCAGATGGCCTCTGGGAAC |
(Pötzsch et al., 2021) | N/A |
| Hcar1_genotyping_fw: AGTTTCTCTCGTGGGTGCAG | (Pötzsch et al., 2021) | N/A |
| Hcar1_genotyping_rev: TGCTTGCCTGGCTCGAATAA | (Pötzsch et al., 2021) | N/A |
| Atf3_sgRNA_fw: CACCGGCGGTCGCACTGACTTCTG |
This paper | N/A |
| Atf3_sgRNA_rev: AAACCAGAAGTCAGTGCGACCGCC |
This paper | N/A |
| Atf3_genotyping_fw: GTGGGCCAGAACATAAAGGA |
This paper | N/A |
| Atf3_genotyping_rev: CCAGAGCTTTGGATGGTTCT | This paper | N/A |
| U6_sequencing: GAGGGCCTATTTCCCATGATTCC |
(Ran et al., 2013) | N/A |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-Puro (PX459) V2.0 | (Ran et al., 2013) | RRID: Addgene_62988 |
| Software and algorithms | ||
| Genome browser (e.g., Ensembl) | http://www.ensembl.org/Mus_musculus/Info/Index | RRID:SCR_013367 |
| CRISPR design tool (e.g., E-CRISP) | http://www.e-crisp.org/E-CRISP/ | RRID:SCR_019088 |
| Other | ||
| Cell culture flasks (T25) | TPP Techno Plastic Products AG | Cat# 90026 |
| Cell culture plates (96-well) | TPP Techno Plastic Products AG | Cat# 92696 |
| Cell culture plates (6-well) | TPP Techno Plastic Products AG | Cat# 92406 |
| Cell culture plates (384-well) | Corning Incorporated | Cat# 353961 |
Materials and equipment
| Proliferation medium | Final concentration | Amount |
|---|---|---|
| Neurobasal medium | n/a | 47.9 mL |
| B-27 (50×) | 1× | 1 mL |
| GlutaMAX (100×) | 1× | 0.5 mL |
| Penicillin/ Streptomycin (10,000 U/mL) | 100 U/mL | 0.5 mL |
| EGF (20 μg/mL) | 20 ng/mL | 50 μL |
| FGF-2 (20 μg/mL) | 20 ng/mL | 50 μL |
| Total | n/a | 50 mL |
Note: Proliferation medium can be stored at 4°C for 2 weeks, but growth factors EGF and FGF-2 should always be added freshly at the time of medium change.
| Wash medium | Final concentration | Amount |
|---|---|---|
| Neurobasal medium | n/a | 48 mL |
| B-27 (50×) | 1× | 1 mL |
| GlutaMAX (100×) | 1× | 0.5 mL |
| Penicillin/ Streptomycin (10,000 U/mL) | 100 U/mL | 0.5 mL |
| Total | n/a | 50 mL |
Note: Wash medium can be stored at 4°C for 2 weeks.
Step-by-step method details
Cloning of sgRNAs into pX459
Timing: 1 week
The vector pX459 can be used to express sgRNA, Cas9 and the selection marker puromycin in the same cell (a map of pX459 is provided at https://www.addgene.org/62988/). The described cloning procedure was modified from (Ran et al., 2013).
-
1.Annealing and phosphorylation of oligonucleotides.
-
a.Reconstitute oligonucleotides in ddH2O to 100 μM.
-
b.Phosphorylate and hybridize reconstituted oligonucleotides.
-
a.
Prepare the following reaction in a 200 μL PCR tube and incubate in a thermocycler for 30 min at 37°C followed by 5 min at 95°C. Ramp down temperature from 95°C to 25°C at 5°C/min.
| Component | Volume (μL) |
|---|---|
| Forward sgRNA oligonucleotide (100 μM) | 1 |
| Reverse sgRNA oligonucleotide (100 μM) | 1 |
| T4 DNA Ligase Buffer (10×) | 1 |
| T4 Polynucleotide Kinase (10 U/μL) | 0.5 |
| ddH2O | 6.5 |
-
2.
Dilute hybridized oligonucleotides 1:250 in ddH2O to obtain a 80 nM sgRNA solution.
-
3.Restriction and ligation into pX459.
-
a.Prepare the following reaction in a 200 μL PCR tube and incubate in a thermocycler for 6 cycles at 37°C for 5 min followed by 23°C for 5 min.
-
a.
| Component | Volume (μL) |
|---|---|
| pX459 (100 ng/μL) | 1 |
| sgRNA solution (80 nM) | 2 |
| Fast Digest Buffer (10×) | 2 |
| DTT (10 mM) | 1 |
| Fast Digest BbsI | 1 |
| T4 DNA Ligase | 0.5 |
| ddH2O | 12.5 |
-
4.Transformation into chemically competent E. coli TOP10.
-
a.Transform 50 μL of competent E. coli with 3 μL of the ligation reaction.
-
b.Follow the heatshock protocol as described in the manufacturer’s manual (https://www.thermofisher.com/order/catalog/product/C404010#/C404010).
-
c.Plate transformed cells onto LB agar plates containing 100 mg/mL ampicillin and incubate for 12–16 h at 37°C.
-
d.Pick a single bacteria colony into 3 mL of LB medium containing 100 mg/mL ampicillin and incubate for 12–16 h at 37°C and 200 rpm.
-
e.Isolate plasmids from 1 mL of the bacteria culture using GeneJET Plasmid Miniprep Kit or other commercial kits.
-
a.
-
5.Validation of successful insertion of sgRNAs into pX459.
-
a.Perform Sanger sequencing using a primer targeting the U6 promoter.
-
b.Align Sanger sequencing trace with the expected sgRNA sequence.
-
a.
-
6.Isolation of endotoxin-free plasmids for transfection.
-
a.Inoculate 250 mL of LB containing 100 mg/mL ampicillin with 0.5 mL of the remaining bacterial culture containing the validated sgRNA plasmids and incubate for 14 h at 37°C and 200 rpm.
-
b.Isolate plasmids using GeneJET Endo-free Plasmid Maxiprep Kit.Alternatives: Other commercial kits and techniques for plasmid isolation are available and can be used. Make sure to include an endotoxin-removal step during plasmid isolation as endotoxins will increase cytotoxicity during transfection.
-
c.Resuspend plasmids in sterile ddH2O and determine DNA concentration.
 Pause point: Plasmids can be frozen and stored until later use.
Pause point: Plasmids can be frozen and stored until later use.
-
a.
Transfection of neural precursor cells and single-cell plating
Timing: 7 days
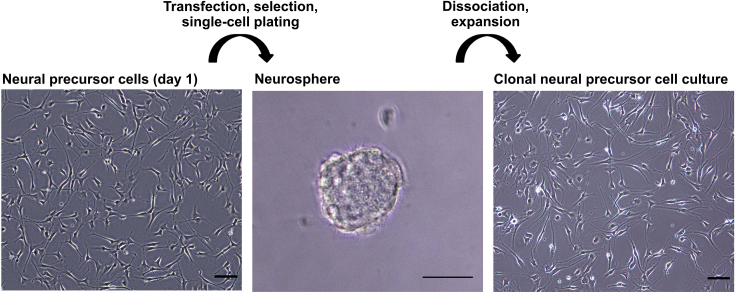
Neural precursor cells are transfected with the respective plasmids pX459+sgRNA. Transfected cells are selected using puromycin treatment and plated at single-cell density to establish clonal cell lines (Figure 2).
Note: This protocol describes the clonal expansion of single neural precursor cells as neurospheres. The outlined steps were optimized for the generation of clonal lines from neural precursor cells that had been isolated from the hippocampus of adult mice. For alternative protocols, which describe the clonal expansion of neural precursor cells derived from other stem cell niches as monolayers, please refer to (Bressan et al., 2017; Dewari et al., 2018).
-
7.Preparation of neural precursor cell cultures.
-
a.Day 1: Passaging of neural precursor cells for transfection. Plate 500,000 cells per T25 flask.
-
b.Culture cells for approximately 48 h until they reach 60% confluence.
-
c.Day 3: Preparation of neural precursor cells for transfection. Perform a half-medium change: remove 3 mL of proliferation medium and replace it with 2.5 mL of fresh proliferation medium.
-
a.
-
8.Transfection of neural precursor cells (day 3).
-
a.Preparation of transfection mix.
-
i.Prepare dilution 1: mix Optimem reduced serum medium with 8 μL P3000 Reagent and 15 μg of plasmid to reach a volume of 250 μL.
-
ii.Prepare dilution 2: mix 240 μL Optimem reduced serum medium with 10 μL Lipofectamin.
-
iii.Add dilution 1 to dilution 2, mix by brief vortexing and incubate for 15 min at 20°C–23°C.
-
i.
-
b.Add transfection mix to cells, swirl plate, and incubate for 36 h in a cell culture incubator.
-
a.
Note: For deletion of gene regions, cells are transfected with two plasmids encoding the different sgRNAs (15 μg each plasmid).
Note: The outlined lipofection protocol causes no visible signs of cytotoxity in mouse hippocampal neural precursor cells and yields transfection efficiencies of 10% with pX459+sgRNA, which is sufficient for generating clonal cell lines.
Alternatives: Other transfection techniques applicable to neural precursor cells, such as nucleofection, magnetofection or nanoparticles, have been described elsewhere (Sapet et al., 2011; Bressan et al., 2017; Wani et al., 2020).
Alternatives: Cas9 and sgRNAs can also be delivered to neural precursor cells by transduction. Make sure to choose the appropriate plasmid for virus-based delivery (https://www.addgene.org/crispr/cut/).
-
9.Selection of transfected cells (day 5).
-
a.Prepare selection medium: add 2 μg/mL puromycin to proliferation medium.
-
b.Replace proliferation medium with selection medium and incubate for 24 h in a cell culture incubator.
-
a.
Note: After incubation with selection medium, a high number of cells are expected to die. The percentage of surviving cells will depend on the transfection efficiency.
-
10.
For recovery of cells, replace selection medium with proliferation medium (day 6).
-
11.Single-cell plating (day 7).
-
a.Detach neural precursor cells from T25 flasks: remove medium, wash once with PBS and incubate with Accutase (0.5 mL/T25) for 5 min at 37°C (or until cells are detached).
-
b.Resuspend cells in 10 mL of wash medium and pellet for 5 min at 300 × g.
-
c.Remove supernatant, resuspend cells in 10 mL proliferation medium and determine cell concentration using a Neubauer chamber.
-
d.Dilute 500 cells in 30 mL proliferation medium containing 2 μg/mL heparin. This corresponds to an approximate cell dilution of 16.7 cells/mL. Mix cell suspension and distribute into an uncoated 384-well-plate (75 μL/well).
-
e.To obtain sufficient numbers of neurospheres, we recommend to plate three 384-well-plates per sgRNA construct.
-
f.Incubate for 14 days in a cell culture incubator to allow neurosphere formation.
-
a.
Note: Addition of heparin stabilizes growth factors such that medium changes are not necessary during the incubation period.
CRITICAL: Proper quantification and dilution of cells is critical. Too high cell concentrations increase the risk of obtaining mixed clonal lines. Too low cell concentrations reduce the numbers of neurospheres obtained.
Optional: After single-cell plating, the cells that were not plated can be frozen or expanded as a mixed population and used to assess gene editing efficiency. Editing efficiency can be tested using methods such as the SURVEYOR assay described in detail in Ran et al. (2013) or by PCR and sequencing. Please refer to the section “Validation of gene knockouts” below for a description of how to detect gene editing events by PCR and sequencing.
Figure 2.
Generation of clonal neural precursor cell cultures
Depicted are bright field microscopy images of the neural precursor cell culture before gene editing (left), a neurosphere formed after plating of transfected cells (middle), and a neural precursor cell culture expanded from a single neurosphere (right). Scale bars: 50 μm.
Expansion of clonal neural precursor cell lines
Timing: [2–4 weeks]
Clonal monolayer cultures of hippocampal neural precursor cells can be obtained by dissociating single neurospheres and expanding them as monolayers.
-
12.
Observe all wells of the 384-well-plate and label the wells that contain neurospheres (troubleshooting 1 and troubleshooting 2)
Note: Comparing total numbers and sizes of neurospheres obtained per plate between knockout and control cultures (transfected with sgRNA against lacZ) gives an estimate of the function of the manipulated gene for neural precursor cell proliferation.
-
13.
Transfer individual neurospheres into single wells of a poly-D-lysine/laminin-coated 96-well-plate using a 100 μL-pipet. Dissociate neurosphere in the 96-well by gently pipetting up and down 10 times.
Note: Cells should attach within the next hours and expand as monolayer cultures (troubleshooting 3).
-
14.
When the cells in the 96-wells reach 80% confluence, passage into 6-well-plate.
-
15.
When the cells in 6-well reach 80% confluence, split cells: Freeze 0.5 million cells and resuspend 50,000–100,000 cells in lysis buffer suitable for analysis of genomic DNA (for instance ATL buffer contained in QIAamp DNA Micro Kit).
Note: At this point, cell lines were already expanded and are ready to be used for experiments directly after genotyping. For alternative strategies, in which cell lines are first genotyped and then expanded, please refer to (Bressan et al., 2017; Dewari et al., 2018).
Pause point: Cell lines can be frozen at this step until later use.
Validation of gene knockouts
Timing: [2 days]
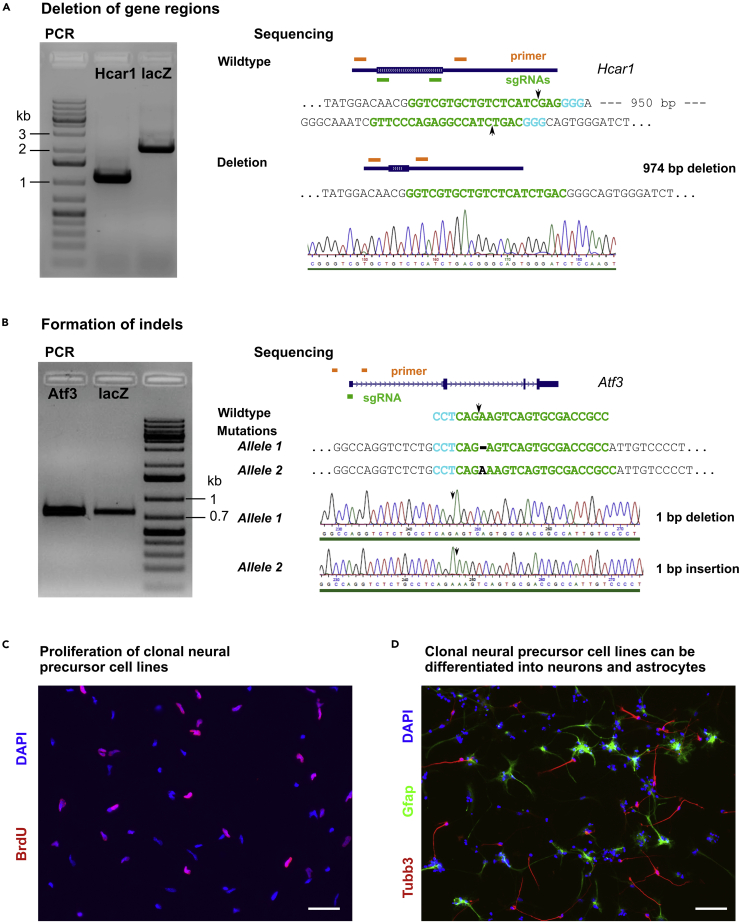
Genotyping of neural precursor cell lines enables the identification and characterization of cultures with homozygous gene knockouts (Figure 3).
-
16.
Isolation of genomic DNA from harvested cells using QIAamp DNA Micro Kit as outlined in the manufacturer’s manual (https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/dna-purification/genomic-dna/qiaamp-dna-kits/) or using other DNA isolation techniques.
Alternatives: Genotyping can also be performed using crude DNA extracts after resuspension and incubation of cells in home-made lysis buffer (Bressan et al., 2017) or commercially available extraction solutions such as QuickExtractTM DNA Extraction Solution.
-
17.
Amplification of genomic sequence flanking the target region using Phusion High-Fidelity DNA Polymerase and the respective genotyping primers.
| Component | Volume (μL) |
|---|---|
| Genomic DNA from cell line (100 ng/μL) | 2 |
| Buffer HF (5×) | 10 |
| dNTPs (10 mM) | 1 |
| Forward primer (10 μM) | 1 |
| Reverse primer (10 μM) | 1 |
| Phusion High-Fidelity DNA Polymerase | 0.5 |
| ddH2O | 34.5 |
| PCR cycling conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial Denaturation | 98°C | 4 min | 1 |
| Denaturation | 98°C | 10 s | 30 cycles |
| Annealing | 60°C | 30 s | |
| Extension | 72°C | 30 s/kb | |
| Final extension | 72°C | 10 min | 1 |
| Hold | 4°C | Forever | |
-
18.
Run 20 μL of each PCR product mixed with 4 μL of 6× loading dye on an 1% agarose gel.
-
19.
Analyze gel and confirm single bands.
Note: Deletions of larger gene regions using the dual sgRNA approach can be detected by a shift in band size of the PCR product. Homozygous deletions should exhibit single bands at the size of the expected mutated fragment and no PCR product at the size of the wildtype fragment (Figure 3A). PCR products at the size of the wildtype fragment can indicate either wildtype alleles or indel formation (e.g., after cutting at the target sequence of only one of the sgRNAs). Since indels can lead to loss-of-function mutations, identification of heterozygous gene knockouts requires sequencing of the PCR product.
CRITICAL: Knockouts using the single sgRNA approach (formation of indels) cannot be detected by PCR but must be verified by sequencing (Figure 3B). Homozygous knockouts must contain indels that result in frameshifts in both of the two alleles. Heterozygous knockouts must contain one wild-type allele and one allele with frameshift mutation.
-
20.
Purify the remaining PCR product (which was not loaded onto the gel) using QIAGEN PCR purification kit or other techniques.
-
21.
Sanger sequencing of the PCR product and alignment of the obtained sequence to the reference sequence of the respective mouse gene.
Note: To obtain sequences of both alleles, mixed Sanger sequencing traces can be deconvoluted using online tools such as TIDE (http://shinyapps.datacurators.nl/tide/), CRISP-ID (http://crispid.gbiomed.kuleuven.be/) or other deconvolution techniques. Please refer to the respective webpages for detailed instructions.
Alternatives: Parallel sequencing of the PCR product (MiSeq) enables quick and accurate detection of indel formation in both alleles and does not require deconvolution of a mixed trace (troubleshooting 4 and troubleshooting 5)
Note: Knockouts of coding genes should be validated at the protein level using western blot or immunocytochemistry with protein-specific antibodies.
Figure 3.
Identification of homozygous knockout lines and representative analysis
(A) Left, PCR-based amplification of the Hcar1 gene revealed a single product at the size of the shortened fragment in a Hcar1-modified clonal culture, suggesting that this culture contains a homozygous deletion. A product at the size of the wildtype fragment was observed in a control (lacZ-transfected) clonal culture. Right, Sanger sequencing of the purified PCR product from the Hcar1-modified clonal culture confirmed the deletion. Arrows indicate cutting site of Cas9.
(B) Sequencing of the PCR product from the Atf3-modified clonal culture reveals a deletion of a single adenosine residue in allele 1 and an insertion of a single adenosine residue in allele 2. Both indels resulted in frameshifts, identifying this culture as a homozygous knockout of Atf3.
(C and D) Representative analysis of knockout neural precursor cells. Proliferation of the modified cell lines can be assessed by BrdU assays as described in (Bernas et al., 2017). (D) The role of the modified genes in formation of neurons (Tubb3-positive) and astrocytes (Gfap-positive) can be analyzed by immunocytochemistry after differentiation of neural precursor cells as described in (Bernas et al., 2017). Scale bars in (C and D): 50 μm.
Expected outcomes
The described protocol allows the generation of clonal mouse hippocampal neural precursor cell lines containing gene knockouts within a time period of seven weeks. The knockout efficiency will vary depending on the genomic location targeted and the cutting efficacy of the sgRNA. Using the single sgRNA approach with the depicted sgRNA targeting Atf3, we observed that 75% of clonal cultures contained homozygous frameshift mutations and 25% of cultures contained frameshift mutations on only one allele. Using the dual sgRNA approach with the depicted sgRNAs targeting Hcar1, we found that 45% of clonal cultures contained homozygous deletions of the region flanked by the sgRNAs.
Clonal neural precursor cell lines without gene knockouts (e.g., sgRNA against lacZ) proliferate at a rate comparable to the original culture (Figure 3C) and can be differentiated into astrocytes and neurons (Figure 3D). Independent clonal cultures should be used as replicates for follow-up analyses, such as for the assessment of the gene knockouts on differentiation potential or molecular pathways.
Limitations
Since the expansion of the neural precursor cell lines is dependent on active cell proliferation, the protocol described here is not suitable for generating cell lines with knockouts of genes that are required for cell division or viability and maintenance of neural precursor cells. Deletion of such genes will lead to the generation of low numbers of neurospheres and, if those can be expanded, to a high percentage of cultures with heterozygous mutations or no mutations. Function of genes required for proliferation can alternatively be assessed in the mixed population acutely after transfection of neural precursor cells without the generation of clonal cell lines.
The expansion of clonal hippocampal neural precursor cell lines as described in this protocol is laborious and takes on average four weeks. Therefore, the procedure outlined here is not suitable for high-throughput screening of gene panels but rather for the in-depth analysis of gene candidates.
One potential limitation of this protocol could be the clonal expansion of cells as neurospheres. First, the numbers of generated clonal cultures will be dependent on the numbers of neurospheres obtained (we obtained approximately 30 neurospheres from each 384-well plate). However, neurospheres can be easily detected and collected from well plates and plating three 384-well plates (as recommended in the protocol) will certainly yield sufficient numbers of clones. A disadvantage of neurospheres is that cells inside the sphere are exposed to lower growth factor concentrations compared to cells at the outside of the sphere, which could lead to cell differentiation inside the sphere. However, this does not reduce the efficiency of generating cultures, since, from our experience, most neurospheres that were formed could be rapidly and reliably expanded as monolayer cultures without signs of cell death.
Troubleshooting
Problem 1
No neurospheres were obtained after single cell plating, also not from control cultures that were transfected with lacZ sgRNA (step 12).
Potential solution
Accurate cell counting before single-cell plating is critical. Too few plated cells will lead to very few neurospheres. We recommend performing cell quantifications in triplicates.
Problem 2
Neurospheres were obtained from control cultures but not from cultures that were transfected with sgRNAs targeting a specific mouse gene (step 12).
Potential solution
The targeted gene might be required for proliferation of neural precursor cells and its knockout might prevent clonal expansion of the cell lines. As an alternative, proliferation and differentiation of neural precursor cells could be analyzed after transfection with pX459+sgRNA in the mixed cell population without clonal expansion (validate that the gene editing efficiency in the mixed population is sufficiently high).
Problem 3
Cells did not attach on the poly-D-lysine/laminin-coated 96-well-plate within 24 h after suspension (step 13).
Potential solution
Forceful dissociation might lead to cell death. Gently dissociate neurospheres and avoid foaming.
Problem 4
Sequence reveals that more than two different indels are present within a single culture (step 21).
Potential solution
Plating of too many cells will lead to mixed cultures during neurosphere formation. Accurate cell counting is critical. We recommend performing cell quantifications in triplicates. Alternatively, mixed lines could be re-plated to derive clonal cultures.
Problem 5
No clonal cultures with homozygous gene knockouts were identified (step 21).
Potential solution
Confirm gene editing efficiency in the mixed culture. If gene editing efficiency is high, the mixed culture could be re-plated at single-cell density to derive more clones. If gene editing efficiency is low, consider choosing a different sgRNA for that particular gene.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gerd Kempermann (gerd.kempermann@dzne.de).
Materials availability
No new materials were generated in this study.
Data and code availability
This study did not generate new datasets or code.
Acknowledgments
This study was financed by basic institutional funds from the Helmholtz Association and TU Dresden. We thank Martin Schneider and Aksana Schneider for their expert advice and for sharing materials. We further thank Rupert Overall for helpful comments on the manuscript.
Author contributions
S.Z. designed the protocol, performed the experiments, and wrote the manuscript. G.K. supervised the work and acquired funding.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Sara Zocher, Email: sarazocher@neuro.mpg.de.
Gerd Kempermann, Email: gerd.kempermann@dzne.de.
References
- Babu H., Claasen J.H., Kannan S., Rünker A.E., Palmer T., Kempermann G. A protocol for isolation and enriched monolayer cultivation of neural precursor cells from mouse dentate gyrus. Front. Neurosci. Front. 2011;5:89. doi: 10.3389/fnins.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernas S., Leiter O., Walker T.L., Kempermann G. Isolation, culture and differentiation of adult hippocampal precursor cells. Bio-Protoc. 2017;7:1–17. doi: 10.21769/BioProtoc.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan R.B., Dewari P.S., Kalantzaki M., Gangoso E., Matjusaitis M., Garcia-Diaz C., Blin C., Grant V., Bulstrode H., Gogolok S. Efficient CRISPR/cas9-assisted gene targeting enables rapid and precise genetic manipulation of mammalian neural stem cells. Development. 2017;144:635–648. doi: 10.1242/dev.140855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Xu J., Cheng M., Liao X., Peng S. Interdisciplinary Sciences: Computational Life Sciences. Springer Berlin Heidelberg; 2018. Review of CRISPR/Cas9 sgRNA design tools; pp. 455–465. [DOI] [PubMed] [Google Scholar]
- Dever D.P., Scharenberg S.G., Camarena J., Kildebeck E.J., Clark J.T., Martin R.M., Bak R.O., Tang Y., Dohse M., Birgmeier J.A. CRISPR/Cas9 genome engineering in engraftable human brain-derived neural stem cells. iScience. 2019;15:524–535. doi: 10.1016/j.isci.2019.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewari P.S., Southgate B., Mccarten K., Monogarov G., O'Duibhir E., Quinn N., Tyrer A., Leitner M.C., Plumb C., Kalantzaki M. An efficient and scalable pipeline for epitope tagging in mammalian stem cells using Cas9 ribonucleoprotein. eLife. 2018;7:e35069. doi: 10.7554/eLife.35069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pötzsch A., Zocher S., Bernas S.N., Leiter O., Rünker A.E., Kempermann G. L-lactate exerts a pro-proliferative effect on adult hippocampal precursor cells in vitro. iScience. 2021;24:102126. doi: 10.1016/j.isci.2021.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapet C., Laurent N., de Chevigny A., Le Gourrierec L., Bertosio E., Zelphati O., Béclin C. High transfection efficiency of neural stem cells with magnetofection. BioTechniques. 2011;50:187–189. doi: 10.2144/000113628. [DOI] [PubMed] [Google Scholar]
- Walker T.L., Kempermann G. One mouse, two cultures: Isolation and culture of adult neural stem cells from the two neurogenic zones of individual mice. J. Vis. Exp. 2014 doi: 10.3791/51225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani T.U., Sofi H.S., Khan N.A., Sheikh F.A. Experimental protocol for induction of transgene expression in neural stem cells through polymeric nanoparticles. Methods Mol. Biol. 2020;2125:77–84. doi: 10.1007/7651_2019_256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new datasets or code.