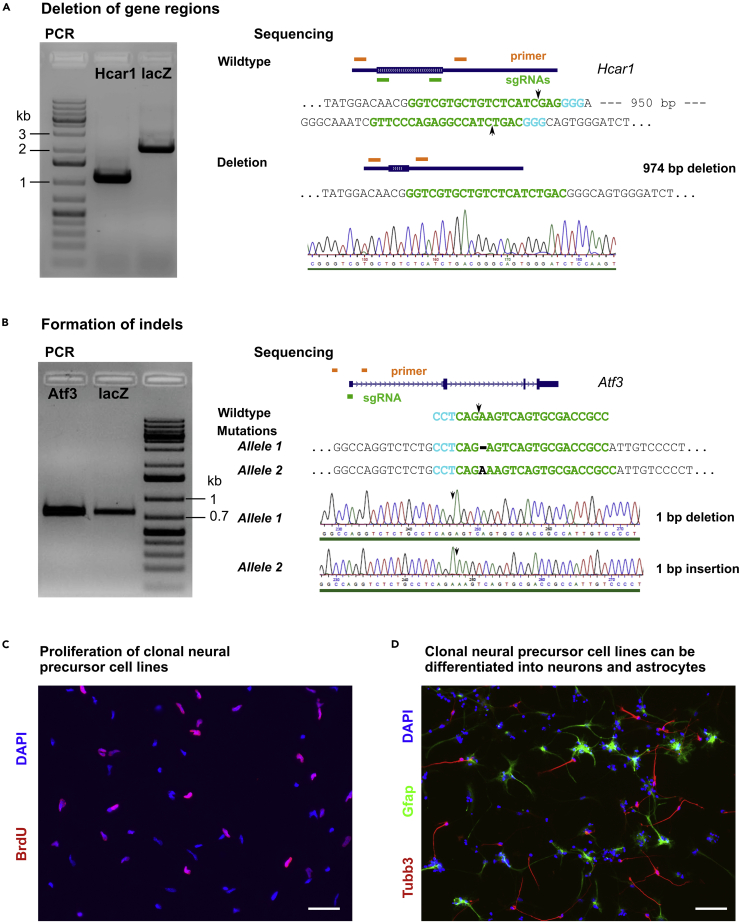
Figure 3.
Identification of homozygous knockout lines and representative analysis
(A) Left, PCR-based amplification of the Hcar1 gene revealed a single product at the size of the shortened fragment in a Hcar1-modified clonal culture, suggesting that this culture contains a homozygous deletion. A product at the size of the wildtype fragment was observed in a control (lacZ-transfected) clonal culture. Right, Sanger sequencing of the purified PCR product from the Hcar1-modified clonal culture confirmed the deletion. Arrows indicate cutting site of Cas9.
(B) Sequencing of the PCR product from the Atf3-modified clonal culture reveals a deletion of a single adenosine residue in allele 1 and an insertion of a single adenosine residue in allele 2. Both indels resulted in frameshifts, identifying this culture as a homozygous knockout of Atf3.
(C and D) Representative analysis of knockout neural precursor cells. Proliferation of the modified cell lines can be assessed by BrdU assays as described in (Bernas et al., 2017). (D) The role of the modified genes in formation of neurons (Tubb3-positive) and astrocytes (Gfap-positive) can be analyzed by immunocytochemistry after differentiation of neural precursor cells as described in (Bernas et al., 2017). Scale bars in (C and D): 50 μm.