Figure 1.
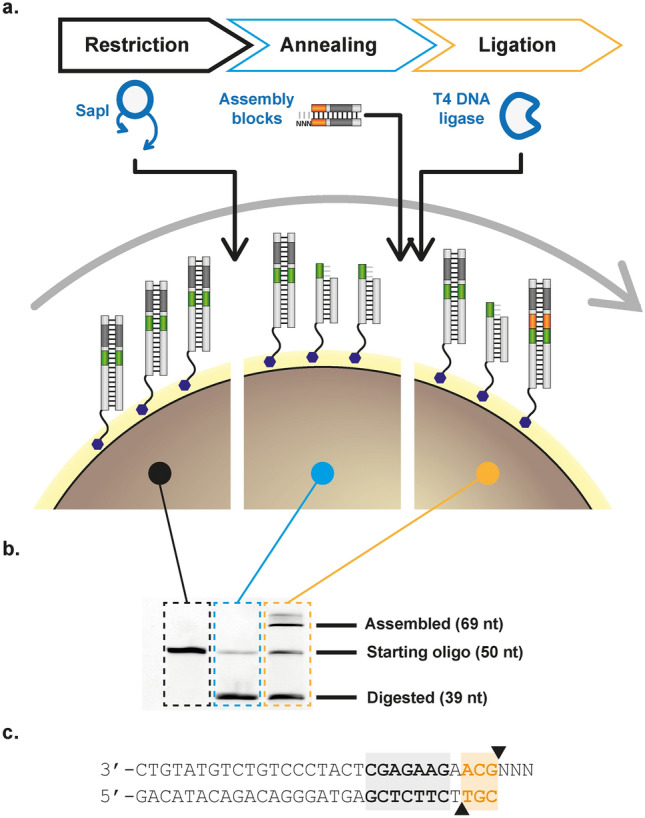
Producing length and compositional variation with InDel assembly. (a) At each assembly cycle, dsDNA templates bound to the paramagnetic beads are restricted with SapI (a type IIs endonuclease), building blocks annealed and ligated. After ligation, the cycle can be restarted. Compositional variation is achieved primarily by combining pools of different building blocks. (b) Denaturing gel electrophoresis of fluorescently labelled template across the different steps of the cycle show that restriction digestion and ligation are not carried out to completion in any step, underpinning the length variation of the resulting libraries. (c) Sequence of a building block. A long double-stranded region is used to stabilize building block annealing and ensure efficient endonuclease activity. SapI recognition site is shown in grey and restriction sites as black triangles. The overhang depicted would add GCA, coding for alanine to the growing chain—further information in Supplementary Table 3.
