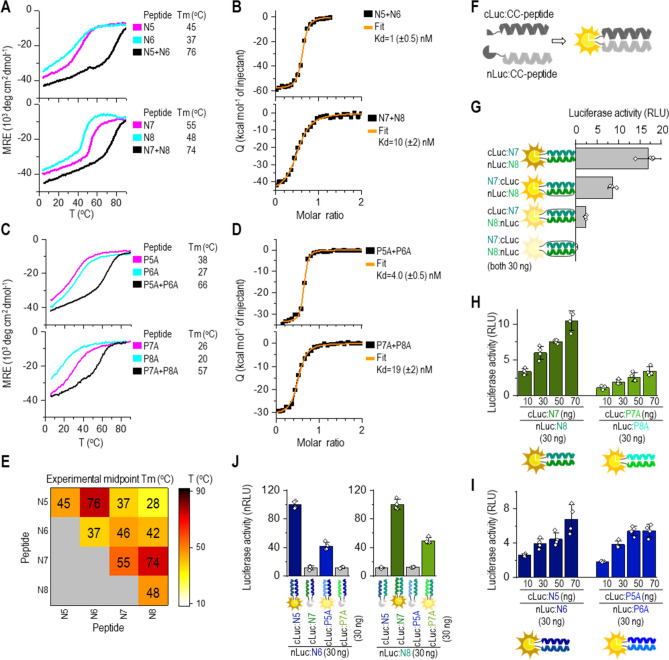
Figure 1.
Orthogonal CC peptide pairs with Kd in the nanomolar range based on modifications at the b, c, and f sites. (A, C) Thermal denaturation profiles of peptides (40 µM; magenta and cyan) and CCs (20 µM each peptide; black) monitored by a CD signal at 222 nm. The midpoint Tm was calculated based on thermodynamic model fit35. (B, D) Isothermal titration calorimetry (ITC) analysis of the binding affinity of designated CC peptide pairs. The binding isotherms of heat release per injection are depicted as a function of the increasing peptide-to-peptide molar ratio. The dissociation constant, KdITC, was calculated using the two-state dimer association model. (E) Heat map of the matrix of the calculated midpoint Tm from thermal denaturation scans of all peptide combinations. (F) Scheme of reconstitution of CC-split luciferase managed by a CC-forming peptide pair. (G) Luciferase activity of reconstituted CC-split luciferase in HEK293T measured 48 h after transfection of HEK293T cells with a plasmid expressing a combination of nLuc tethered to N8 (30 ng) and cLuc tethered to N7 peptide (30 ng). (H, I) Luciferase activity determined 48 h after transformation of HEK293T cells with plasmids expressing nLuc:N8 or nluc:N6 (30 ng); and cLuc tethered to N7 or P7A or cLuc tethered to N5 and P5A (10–70 ng). (J) Orthogonality of designed N peptide set in HEK293T cells by co-transfection of the nLuc:N8 or nLuc:N6 fusion encoding plasmid (30 ng), and cLuc:CC (CC stands for N5, P5A, N7, P7A) (30 ng). Reconstituted luciferase activity was measured 48 h after transfection. Amounts of plasmids are indicated in Table S2. The values (G-J) represent the means (± s.d.) from four independent cell cultures, individually transfected with the same mixture of plasmids, and are representative of two independent experiments.