Fig. 4. Distinguishing neoplastic and nonneoplastic cells.
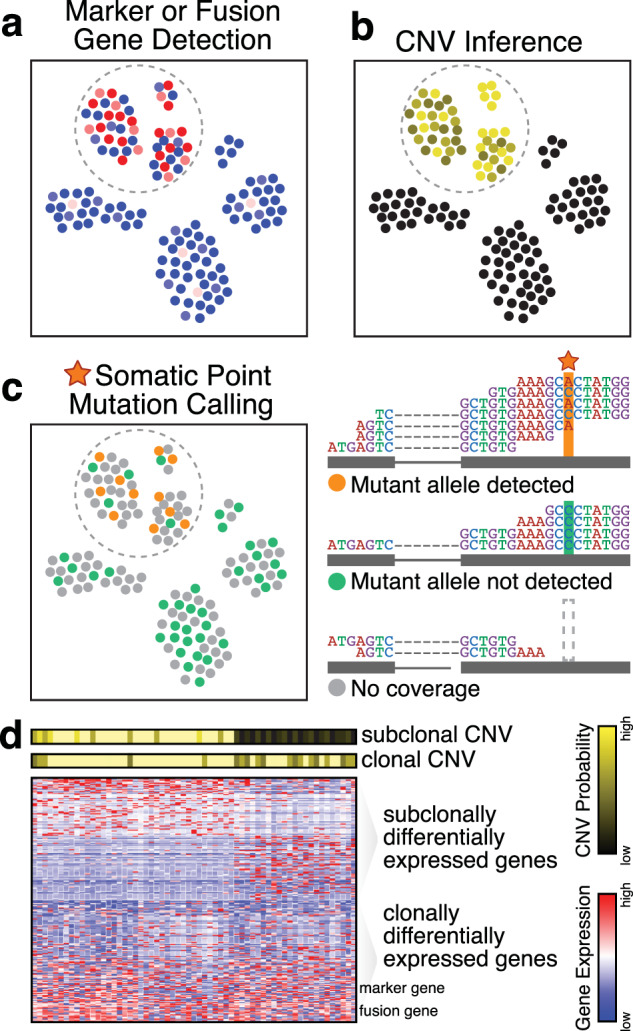
a The detection of marker or fusion genes that are uniquely upregulated or expressed in neoplastic cells may be used to identify neoplastic cells. In this illustration, many neoplastic cells exhibit high expression (red) of a marker or fusion gene, although dropouts or other technical factors result in the detection of low or no expression (blue) in other neoplastic cells in the same cluster. b Copy-number variant (CNV) inference may also be used to identify neoplastic cells. Normalized smoothed gene expression magnitudes and variant allele frequencies can be used to infer the probability that a cell harbors CNVs. Neoplastic cells exhibit higher probabilities of harboring any CNVs, as expected. c Somatic point mutation calling may be used to identify neoplastic cells. The top read pileup for a cell shows an example in which both the mutant and reference alleles are detected, indicating that the cells harbors the mutation. The middle read pileup for a cell shows an example in which the mutant allele is not detected, which could indicate that the cell does not harbor the mutation or that there is allelic dropout of the mutant allele. Alternatively, the bottom read pileup shows an example where the mutation site presents no read coverage, and thus, no mutation call can be made. d The inference of CNVs and other genetic alterations directly from RNA-sequencing data enables the direct interrogation of transcriptional differences among genetic subclones. A clonal CNV distinguishes neoplastic from nonneoplastic cells and is also marked by high expression of marker and fusion genes. In addition, a subclonal CNV is present. Differential expression analysis may be applied to directly identify differentially expressed genes between genetic subclones.
