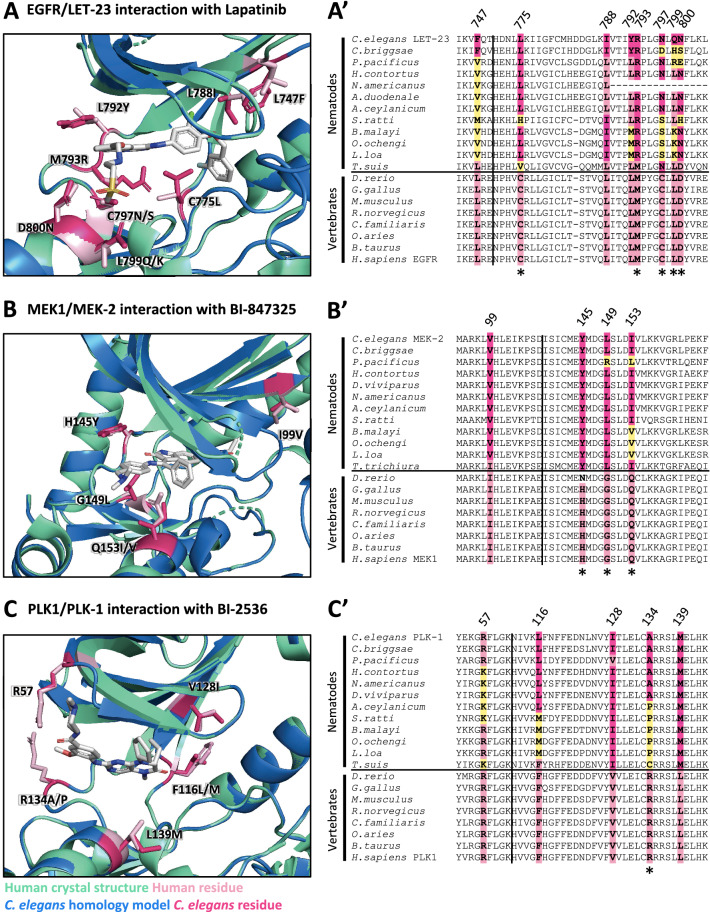
Figure 2.
Structure analysis identifies LET-23, MEK-2 and PLK-1 as candidate targets for anthelmintic development. C. elegans homology models (blue) for LET-23, MEK-2 and PLK-1 aligned with human crystal structure (green) for EGFR (PDB: 1XKK)86, MEK1 (PDB: 5EYM)59 and PLK1 (PDB: 2RKU)64 are shown in (A–C) respectively. Key divergent residues proximal to the inhibitor binding sites are highlighted in dark pink (C. elegans residue) and light pink (human residue) within the structure diagrams (A–C). Residues are labeled according to their position in the human kinase, with the first letter indicating the identity of the vertebrate residue and the latter indicating the identity of the nematode residue(s). The conservation of these residues among free-living nematodes, parasitic nematodes and vertebrates is displayed in the corresponding sequence alignments (A′–C′). The vertical line indicates a discontinuous break in the sequence. The horizontal line indicates the separation between the nematode and vertebrate sequences. Yellow residues in the sequence alignments highlight those nematode residues that differ in identity from both vertebrate and C. elegans sequence at the location of these divergent residues of interest. Residues that have distinct physicochemical properties between vertebrates and nematodes are indicated with an asterisk below the alignment.