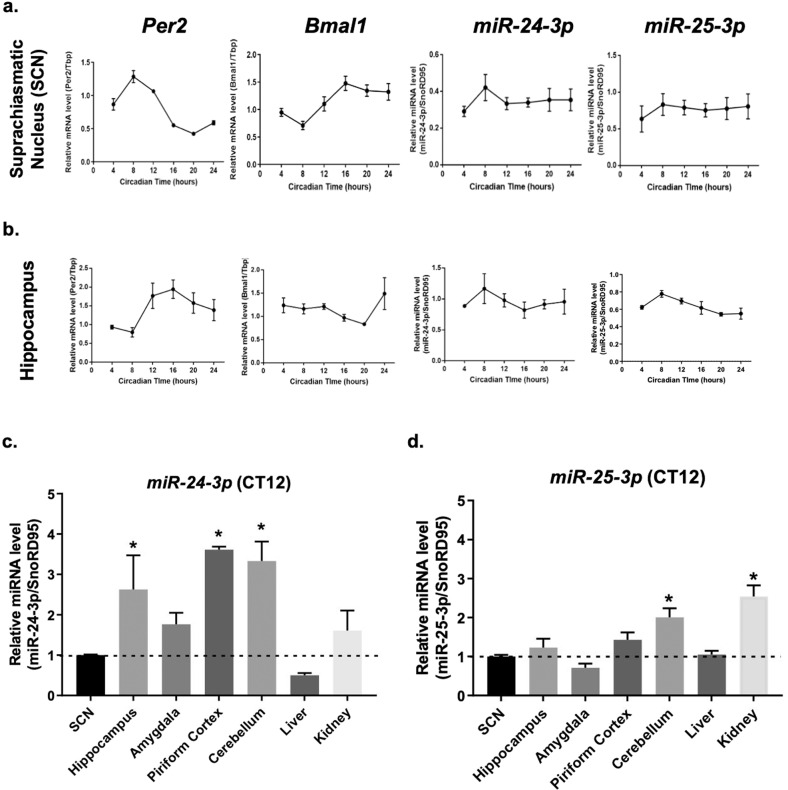
Fig. 6. Expression levels of miR-24-3p and miR-25-3p vary in brain regions and peripheral organs.
WT mice housed under constant dark conditions for seven days were sacrificed at CT04, 08, 12, 16, 20, and 24 h for measurements of the indicated brain and peripheral tissues. Expression profiles of the circadian clock genes (Per2 and Bmal1 mRNAs), miR-24-3p, and miR-25-3p in the a SCN and b hippocampal brain tissues were obtained by real-time qPCR with the relative quantification method. Expression levels of c miR-24-3p and d miR-25-3p in various brain and peripheral tissues were examined. CT12 tissue samples were used and normalized to the SCN miR-24-3p or miR-25-3p level. Data for the Per2 and Bmal1 mRNAs were normalized by the TATA-box binding protein (Tbp) housekeeping gene, while the miR-24-3p and miR-25-3p data were normalized by small nucleolar RNA, C/D Box 95 (SnoRD95). Error bars represent the means ± SE of A.U. for each time point measured in three independent measurements. Significance was assessed by one-way ANOVA, *p < 0.05 compared to the control group.