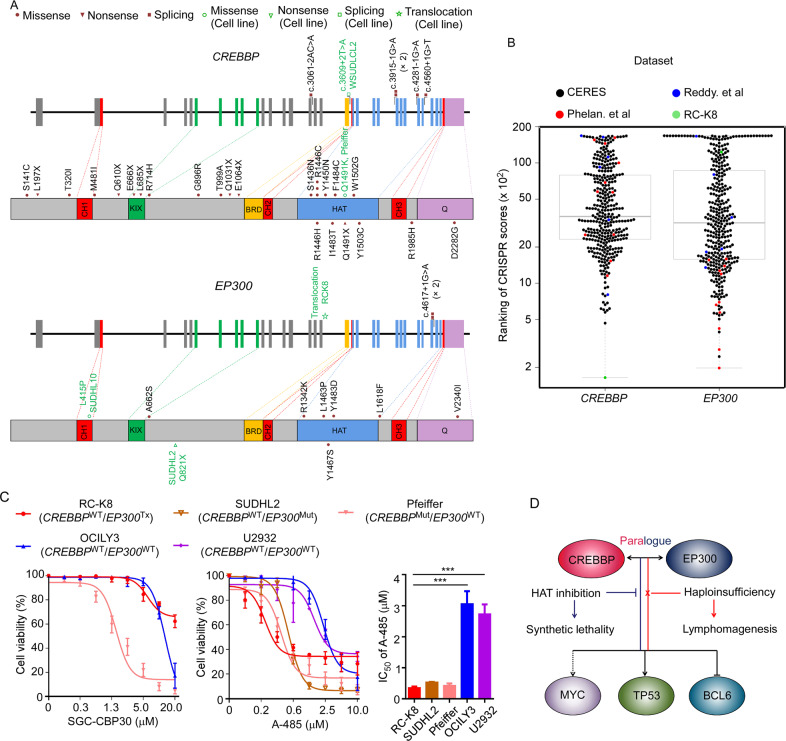
Fig. 5. DLBCL cells with genetic alterations in EP300 and CREBBP are sensitive to a HAT domain inhibitor.
A Somatic mutations in CREBBP and EP300 identified in a previously described DLBCL cohort, RC-K8 cells and other cell lines included by a previous CRISPR screen or tested by A-485 in this study. B Ranking of CRISPR scores of CREBBP and EP300 from different datasets. The middle boxes represent the middle 50% of values for each group with a midline as the median value. The bars outside the boxes represent the 1.5 interquartile ranges outside of the boxes. C Cell viability tests of targeted inhibition of the BRD domain by the small molecule SGC-CBP30 and the HAT domain by A-485. Error bars: SD (n = 3 replicates). The three HAT-mutant cell lines displayed higher sensitivity to A-485 than the cell lines (OCILY3 and U2932) with wild-type HATs, with RC-K8 cells exhibiting the lowest IC50 value. Mut: mutation; Tx: translocation; WT: wild-type. D Potential mechanism of HAT inhibition in HAT-deficient cells: haploinsufficiency of CREBBP or EP300 promotes lymphomagenesis via dysregulation of BCL6 and TP53; the deficiency develops dependency on the remaining HAT function simultaneously. Targeted inhibition of HAT could cause synthetic lethality through abrogation of MYC.